What's New in MacVector 12.5?
Overview
The most important additions to MacVector 12.5 have been a series of improvements to the add-on Assembler module to handle reference alignments of large numbers of next generation sequencing reads using the popular Bowtie assembler algorithm.You can find more details in blog posts here and here and on the Assembler product page.
The main MacVector product has new Multiple Sequence Alignment algorithms (Muscle and T-Coffee), rewritten auxiliary editor windows and numerous enhancements to the single sequence Editor and Map tabs. More details are in the MacVector 12.5 Release Notes.
Multiple Sequence Alignment Algorithms
We have added support for the popular Muscle and T-Coffee multiple sequence alignment algorithms, in addition to the existing ClustalW algorithm. These are accessed simply by choosing the Analyze | Align Multiple Sequences Using | ... sub menu or my clicking on the Align toolbar item and choosing the algorithm from the new popup menu.
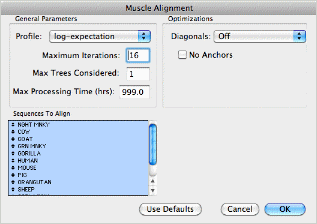 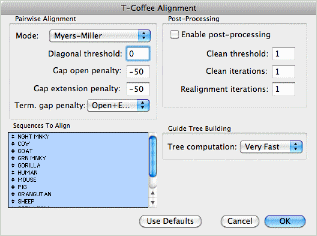
Other enhancements to the multiple sequence alignment functionality include a new scoring function to let you compare alignment methods or evaluate the effects of manual editing and a number of minor improvements to standard OS X scrolling and editing support.
Single Sequence Window Enhancements
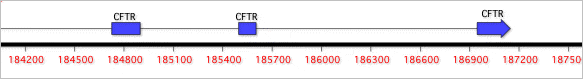
There is now the option to view CDS features as translations in the Editor tab, accessed through the Strands toolbar button. Several new "segmented" feature graphics type have been added to the Map Symbols Options. When these are chosen, segmented features are linked together with lines, making it clear which graphical objects are representing e.g. the individual exons in a segmented CDS.
Miscellaneous Enhancements
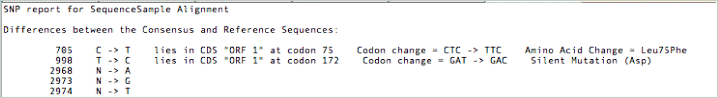
The SNP Reporting in the Align to Reference SNP tab has been enhanced to report differences between the consensus and reference sequences, including any amino acid changes that the change would have on any CDS feature belonging to the reference.
The Restriction Enzyme, Proteolytic Enzyme, Nucleic Acid Subsequence, Protein Subsequence and Scoring Matrix editors have all been recoded using Cocoa. They now follow the standard OS X navigation and selection shortcuts so it is easy to select, copy and paste into different windows to create your own subsets of auxiliary sequence files.
The Align to Reference and Auto-annotation algorithms have had performance enhancements so that they should be noticeably faster than in previous releases.
|

2x.png)


2x.png)
