What's New in MacVector 12.6?
Overview
The most significant addition to MacVector for the 12.6 release is a new interactive "Quicktest Primer" primer design tool that simplifies the identification and testing of primers for PCR and sequencing.
Other enhancements to MacVector include the ability to import features from BED, GFF, GFF3 and GTF files, improvements in graphics performance for genomes with many features and revised melting temperature calculations to use more recent algorithms. A full list of the changes can be found in the Release Notes.
The Assembler module has also had improvements - in particular, you can now use phrap for de novo assembly of moderate numbers of NGS reads (up to ~500,000 on a typical machine). The Release Notes can be viewed here
Quicktest Primer
There is a new primer design window that displays a variety of information (melting temperature, length, GC composition, homopolymer runs, heteropolymer runs, potential hairpin loops, potential for forming primer dimers or duplexes, thermodynamic properties, absorbance etc) of any primer entered in the main edit box;
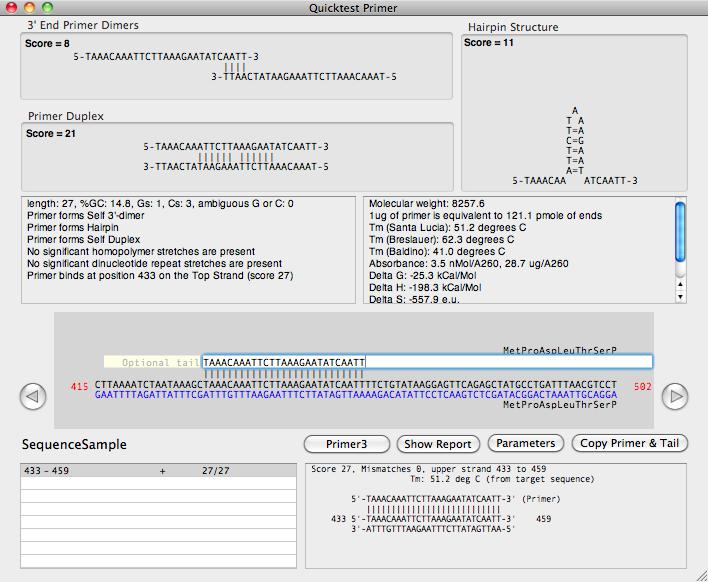
The main editor pane also shows the primary binding site of the primer on the currently active sequence window (if one is open). Any secondary binding sites are shown in a scrolling list and can be clicked on and directly viewed in a separate alignment panel.
The window is highly interactive - if you bring another sequence window to the front, it will instantly update to display the potential binding sites of the primer (if any) on that sequence. You can use the arrow buttons to "nudge" the primer along the sequence to try to reduce potential hairpin or primer dimer/duplex formation. The translations of any CDS features in the sequence are displayed and you can edit the primer sequence to introduce mismatch mutations and any amino acid changes that would result are instantly displayed;
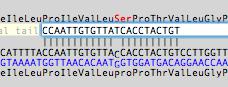
You can also add a tail to the primer (e.g. containing a restriction site) in the "Optional Tail" box. All of the Tm, hairpin/dimer/duplex, absorbance, molecular weight etc calculations then take the tail into account, but the reporting of template binding sites is unaffected.
A useful "Show Report" button opens a window summarizing all of the properties on a single PDF page, great for printing out and inserting into a lab notebook.
Once you are satisfied with a primer, you can click on the "Primer3" button to find a matching primer for a PCR reaction. The Primer3 interface has been modified so that it understands any tails that have been added. You can add tails to any matching primers found by Primer3 and then copy the predicted product to create a new sequence, complete with mismatches and tails, representing the fragment produced by the PCR experiment.
For more details on how to use this exciting new functionality, take ten minutes to look at the Primer Design Tutorial
|

2x.png)


2x.png)
