This is part of a series of posts about and leading up to the release of MacVector 12.5.
With Assembler 12.5 our developers have come up with an affordable and straightforward solution for assembling and visualizing your NGS data. Generating sequencing data is cheaper than it has ever been, however, with this increase in data has come a problem with analysis. Assembler will now create reference assemblies with just a few mouse clicks using Bowtie. Instead of sending your millions of reads away to be assembled or delving into complicated software tools you’ll be able to align millions of NGS reads to multi megabase reference sequences in literally minutes. Bowtie is a fast algorithm, and although it’s an ungapped assembler, what it loses in accuracy it makes up for in speed. You do not need a 32GB 8core Mac Pro to assemble your data. In addition to the existing phrap/phred tools this makes Assembler a simple, cost-effective solution to analyzing your Next Generation Sequencing reads.
Creating a Reference Assembly
- – Chose File | New | Assembly Project to create a new empty project file.
- – Click on the Add Reads tool bar button, then select the sequence files you wish to assemble and click on the Open button. Read(s) file(s) can also be drag and dropped on the open Assembly Project window.
- – Click on the Add Ref tool bar button, then select the sequence file you wish to align the reads against and click on the Open button.
- – Choose Analyze | Bowtie to run the Bowtie algorithm on all of the sequences in the project. Note that if no sequences are selected, Bowtie will be run on ALL of the files in the project. However, if any sequences are selected then the reference sequence and at least one reads file must be selected.
Your reference sequence can be in any “openable” format. However, your reads need to be in FASTQ format.
Hit Reporting
In the dialogue you’ll see an important setting called Hit Reporting. Bowtie uses a concept of strata to score alignments. A stratum is defined by all reads that contain the same number of mismatches in the seed (the seed is the first “n” bases of a read which is given higher priority in scoring than the entire read). You can either show ALL ALIGNMENTS, REPORT BEST ALIGNMENT ONLY (show the best alignment in the stratum with the least amount of mismatches) or REPORT ALL BEST ALIGNMENTS (which shows the best alignment in all strata). Which you choose depends on a few factors. For example how many references you have, how many repeated regions you expect, whether you are using a reference sequence from the same organism or a related one, and many others. Generally start with show all alignments, which is the quickest, and work from there.
Analysis
..and that’s how easy it is. Of course generating results is always easier than analysing them and to help analyse your reference contig Assembler has a few useful tools. We’ll talk about variant detection in a later blog post, but the coverage map is one of the first tools that you will see upon completing an assembly.
Using the Coverage Map
It is extremely useful to be know the depth of reads that are aligned on your reference. Areas of low coverage indicate that you need further sequencing and peaks of high coverage can be indicative of repeats. The Map view of a reference contig will show details of the depth of reads in a coverage map with four statistics. A single plot line shows a running average of the number of reads at that point. However, an average plot is not very sensitive when viewed at a high level and so two shaded areas indicate the maximum value and the minimum value of the averaged reads at that point. As the coverage map is viewed at a lower level these three values will become increasingly closer to the extent that when viewed at, or close to, residue level these three plots will become identical. Areas of zero coverage are shown in light grey. Note that these areas are always displayed even when they are disproportionate to the level of magnification.
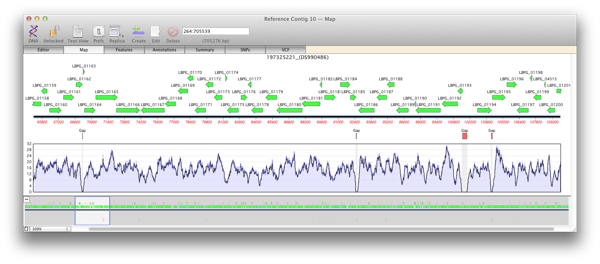
Multiple reference sequences
You can add multiple reference sequences and depending on the settings reads will be aligned against the best match or against multiple ones. This is great for such tasks as identifying a sequenced isolate amongst a series of closely related strains of virus or bacteria. Having multiple reference sequences helps determine which is most closely related (or identical) to the isolate.
Paired end reads
Paired end reads are very useful for improving the accuracy of alignments and also for indel detection. Paired end reads are created by sequencing both ends of the same DNA molecule, with known fragment size. Since the two reads are now separated by a known distance assembly and orientation of the two reads is less complicated. For Assembler if your reads are paired end all you need to do is ensure that the same filenames but appended with version numbers and Paired End assembly is enabled.
e.g.
READS_1.fastq READS_2.fastq
You’ll also need to input the fragment size.
In the next Assembler post we’ll talk about variant detection.
..and remember if you purchase an upgrade or a new license before the release of MacVector 12.5 you can get Assembler with a 50% discount and a free upgrade to MacVector 12.5 when it is released. This offer ends on 1st December. Please request a quote now. Don’t forget to quote the promotional code of “Assembler50%”
