While most MacVector analysis algorithms can handle circular sequences, its sometimes useful to be able to set the “12 o’clock” point to a different location on the circle. This is easy to do with recent MacVector releases.
If you are using MacVector 13.5, then all you have to do is position the flashing caret between the residues where you wish the new origin to be and then right-click (or ctrl-click if you don’t have a right mouse button) and select Set Circular Origin from the context-sensitive menu that appears;
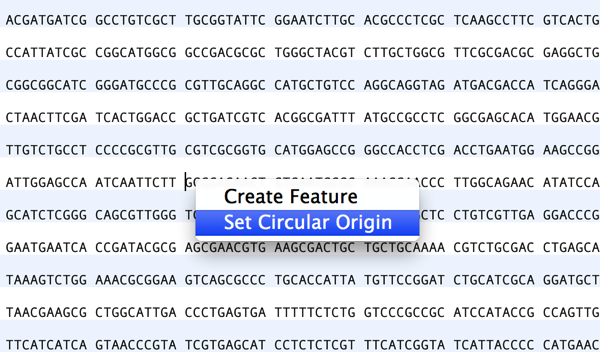
If you have an earlier version of MacVector (12.7 or later), you can use the Cloning Clipboard to reset the circular origin. This does require that there is a suitable restriction site at the location where you wish the new origin to be. Open the circular sequence, switch to the Map tab, click on any restriction enzyme site, then click on the Digest toolbar button.
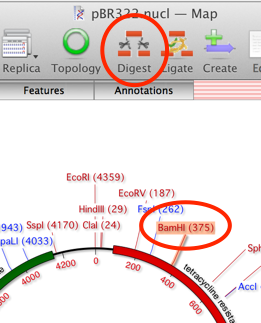
That will place the linearized vector onto the Cloning Clipboard. Now click on the Circularize button on the Cloning Clipboard
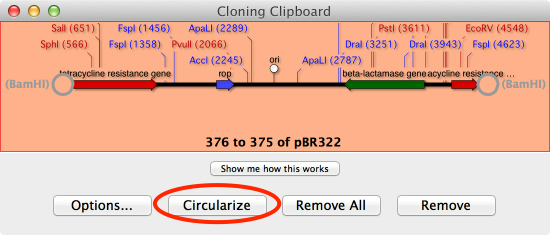
Then click on the Ligate button in the drop down sheet;
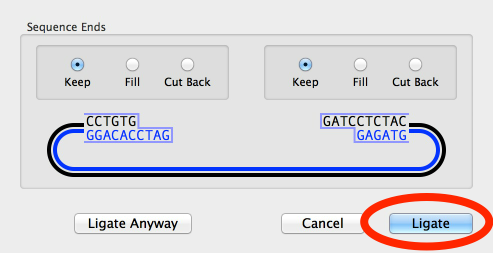
A new circular DNA sequence will be created with the selected restriction enzyme site now at the 12 o’clock position;
This is an article in a long running series of tips to help you get the most out of MacVector. If you want to get notified every time a new tip gets published, follow us @MacVector on twitter (or check the feed for the hashtag #101MacVectorTips) or like us on Facebook.