MacVector currently does not have a built-in function for recognizing restriction sites that are blocked by the DAM or DCM methylases. However, there is a simple workaround that lets you visually identify those sites in the restriction enzyme analysis result windows. The basic idea is that you create additional restriction enzyme sites for each enzyme containing the recognition sequence that is affected by methylation. E.g. ClaI (ATCGAT) will not cleave the sequence ATCGATC. So we can create a DAM-ClaI recognition sequence in a MacVector formatted restriction enzyme file
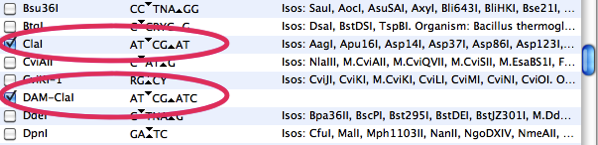
When you search a sequence with this file, you can immediately see which ClaI sites will be blocked by methylation;
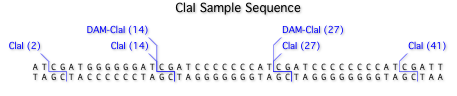
Too tedious to type in all those methylated sites? Not to worry, we’ve done all the hard work for you! You can download a pre-formatted file and copy/paste the full set of DAM-blocked enzymes into your own favorite file. If you are interested in learning more about this, you should definitely check out the complete blog post.