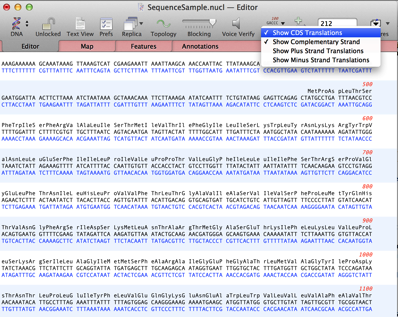
For those of you who like to type in DNA sequences, or do manual editing to the sequences, it’s really useful to be able to see the translations above or below the sequence residues in the Editor tab of the single sequence window. If you are a long term user, you’ll be familiar with the old “Strands” toolbar button that simply toggled on/off the complementary strand. This has changed over recent releases (including a name change to “Display“) to incorporate a variety of different display options.
You can show the 3/6 frame translations by choosing Show Plus Strand Translations or Show Minus Strand Translations. But perhaps the most useful option is the Show CDS Translations item. With this selected only the actual translated amino acid sequences are shown. By “actual”, the display keys off any annotated CDS Features in the Features list. Any sequence you download from Genbank will have CDS features assigned for all of the translated open reading frames. If it’s your own sequence, you should always make a point of assigning a CDS Feature to any segment of DNA that you believe is translated to protein – there are many places in MacVector that specifically key off CDS features, not just this Editor display.