The Database | Align To Folder… function in MacVector is remarkably powerful. Its like having your own personal BLAST search except that it can also scan through millions of Reads in fasta or fastq formatted files to identify those matching an input sequence, which can be DNA OR protein. In addition, it understands about paired-end read files so that when it finds a match, it can retrieve both Reads of a pair even if only one of them matched the input sequence. Lets examine this more closely with a specific example.
Here I am starting off with the protein sequence of a galactokinase enzyme from Streptomyces coelicolor. Lets identify those reads that might encode the protein in an Illumina Solexa dataset from an unknown Clostridium species, retrieve those into a pair of fastq files (one each for the left and right reads) then assemble them to determine the coding region of the protein in the Clostridium sp. This is obviously an artificial example, chosen largely because S. coelicolor DNA is 73% G+C, so there is unlikely to be much sequence similarity between it and a Clostridium species at the DNA level. This illustrates the power of using the Align To Folder function in translated mode to scan for DNA sequences that could encode a specified protein.
First, I open the GalK protein sequence and choose Database|Align To Folder…
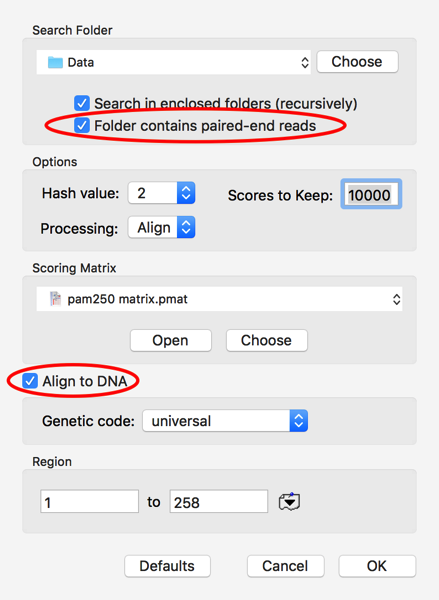
I clicked on the Choose button and selected the folder where my pair of fastq files are located. The other critical settings are to let MacVector know to expect paired files in the folder and, for this example, to check the Align To DNA checkbox. This tells MacVector to translate each DNA Read in all 6 possible frames and compare the translations to the GalK protein. Finally, I set Scores To Keep to 10,000 to be sure I’m keeping all of the potential hits.
To give you an idea of performance, searching this 387aa protein against a total of 19 million x 90nt Reads in two fastq files took a little over 2 hours on a 3 year old 2.7 GHz MacBook Pro. At no time did MacVector use more than 500 MB of RAM, so this analysis can be performed on fairly modest machines, as long as you are patient.
Once complete, you can view a graphical overview of the alignments, a text view showing the actual translated alignments and a list view with each of the “hits” displayed, one per line. The Folder Aligned Sequence view shows that many of the translated reads do indeed show good similarity with the query sequence.
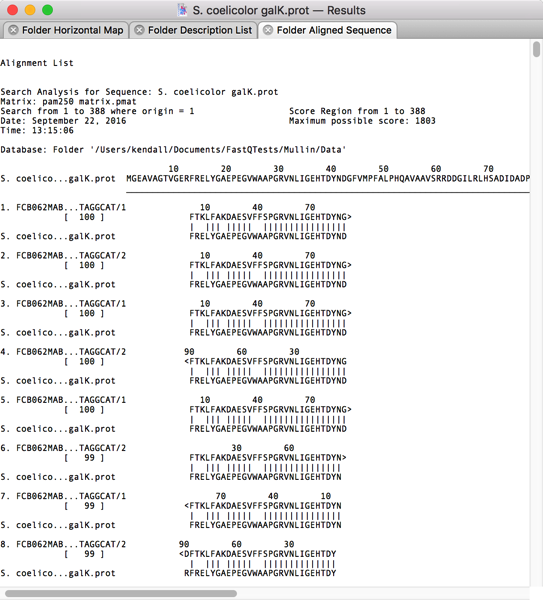
After scrolling through the alignments, I could see that most of the top ~1,000 hits had reasonable similarity to the GalK protein. So I switched to the Folder Description List tab and selected the first 1,000 hits;
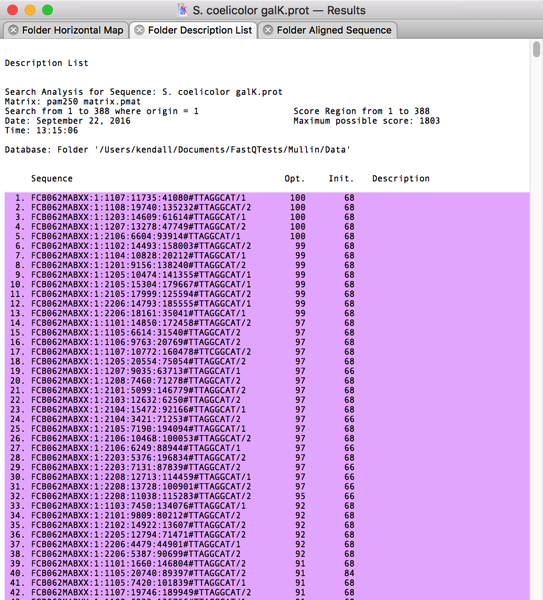
When you select lines containing sequence hits in a Description List like this, A variety of Retrieve To Xxx… items become activated in the Database menu. I selected Database|Retrieve To File… and was prompted for a filename;
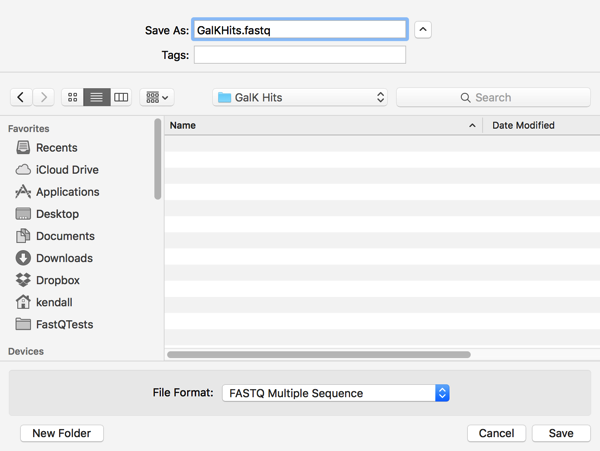
However, once I click on Save and look in the destination folder, I see that two files have been created;
MacVector is smart enough to save BOTH sequences when either one of them is selected in the Description List. Not only that, it keeps them in separate files, so you can use them for further NGS analyses that take paired-end read files, like Bowtie or Velvet for example.
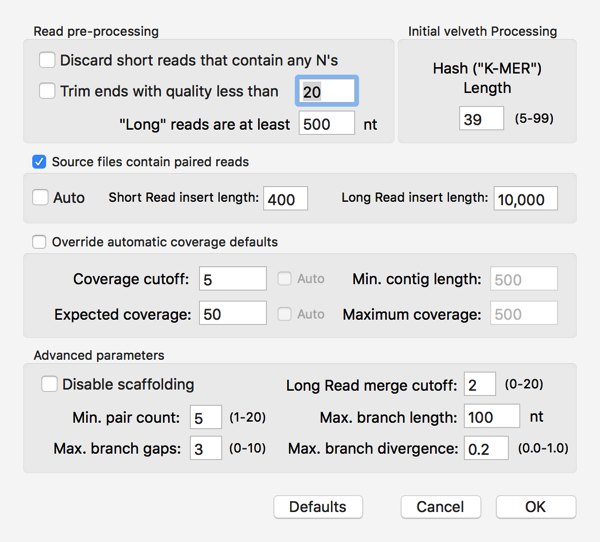
So, can I assemble these into a contig encoding the GalK protein of our Clostridium sp.? I used File|New|Assembly Project to create a new Assembly project then clicked on the Add Seqs toolbar button to add my files. I then clicked on the Velvet toolbar button and accepted the default parameters, except that I checked the Source files contain paired read checkbox;
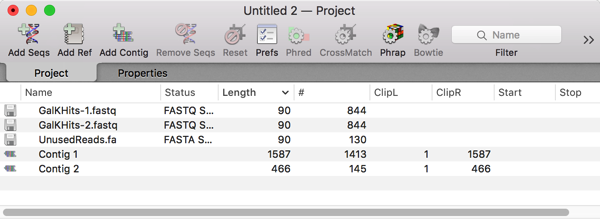
Velvet takes just a few seconds to assemble such a small number of sequences. Encouragingly, there were just two contigs generated, with the longest one (1587bp) containing most of the reads;
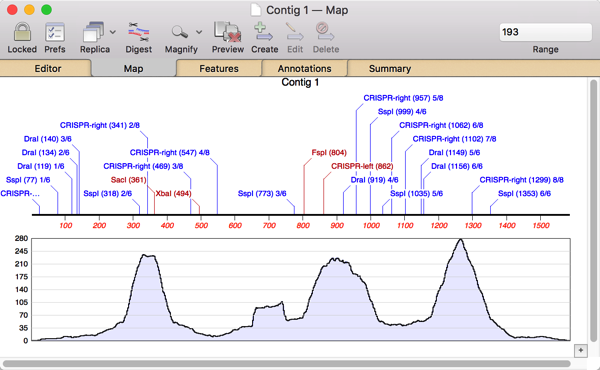
So I double-clicked on Contig 1 to open up the contig editor and switched to the Map tab where I could see the restriction sites and an overview of the assembly;
Perhaps the entire GalK protein is encoded within the Contig? So I chose Analyze|Open Reading Frames…, accepted the defaults and saw one long ORF running from right to left on the minus strand;
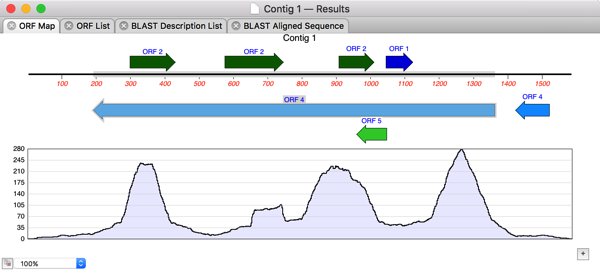
After selecting this, its trivial to choose Analyze|Translation… and get the amino acid translation in a separate MacVector window. Finally, lets run a Database|Internet BLAST search on that translated protein to see what we have “cloned”.
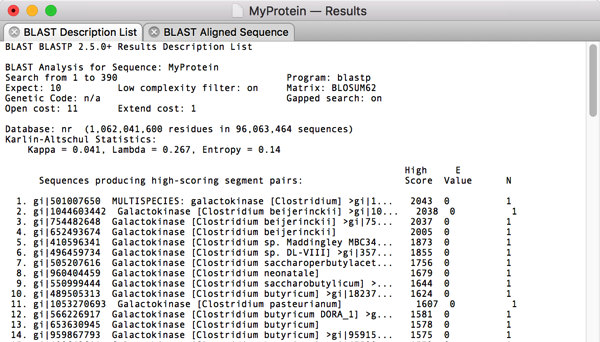
Surprise! The top hit is a Galactokinase protein from another Clostridium species. So there you have it – sorry about the long post but hopefully this gives you a good feeling for how you can use the MacVector Align To Folder function to clone genes from NGS data sets and validate the results.
This is an article in a long running series of tips to help you get the most out of MacVector. If you want to get notified every time a new tip gets published, follow us @MacVector on twitter (or check the feed for the hashtag #101MacVectorTips) or like us on Facebook.
