You can use the Analyze | Open Reading Frames function to very quickly find ORFs on a sequence. Did you know that you can very quickly turn those results into permanent CDS features on your sequence? After running the Open Reading Frames analysis, simply drag and drop the ORF objects you are interested in from the Results window onto the original sequence window. Here we are dropping the results into the Map tab but it works with any of the tabs.
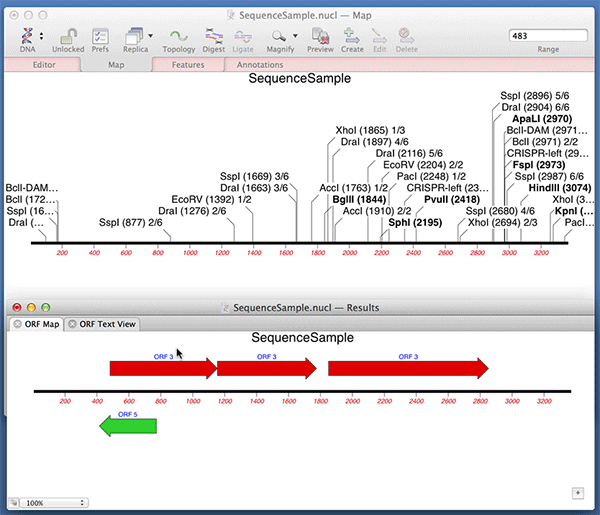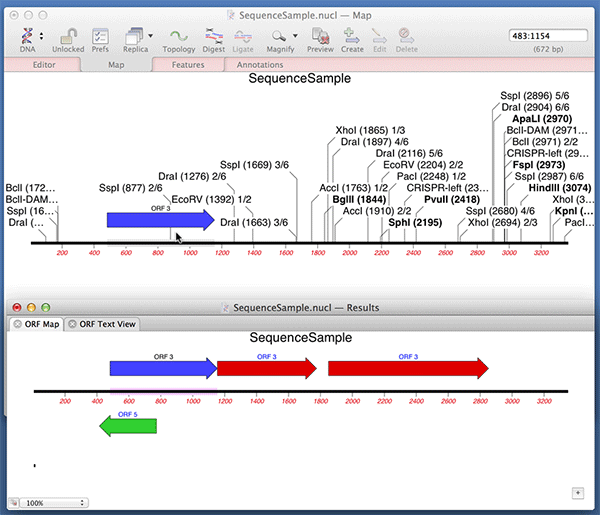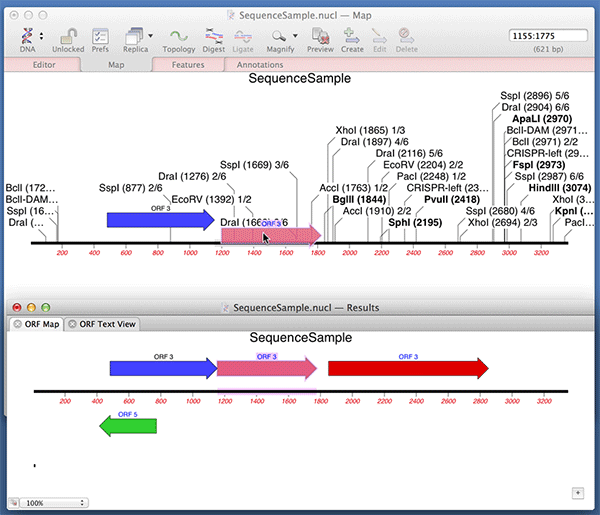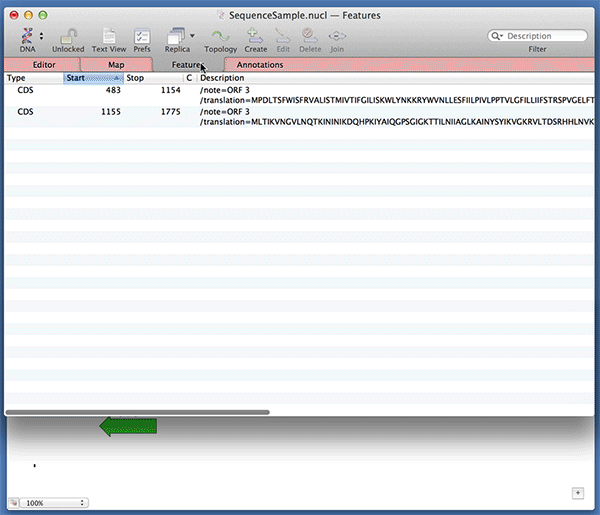
Note that MacVector automatically fills out the /translation= qualifier for you with the predicted amino acid sequence. For optimum Genbank compliance, you should also manually add a /gene= qualifier with the preferred short name of the gene encoding your open reading frame and /product= with the full name of the product..
