When you run a Blast search, as well as a list of hits, you will get a list of alignments between your query sequence and each hit. As with most other text alignments in MacVector, identical matches are by default represented by a vertical line (a score greater than 1) and mismatches (whether similar or not) are represented with a space.
However, sometimes you are more interested in identifying gaps or mismatches in the Blast hits. For example when you are looking for mismatches in motifs or other protein domains or looking for SNPs in DNA sequences.
Most results and displays in MacVector are customizable, and BLAST alignments are no exception. You can also change the length of each line.
To change the match characters:
The default is to display a vertical line for a hit, and a space for mismatches. In the screenshot below we have changed matches to a space, a “-“ for scores between 1 and -1 and “|” for mismatches with a score less than -1. This makes mismatches very noticeable whilst scrolling through the aligned sequence results.
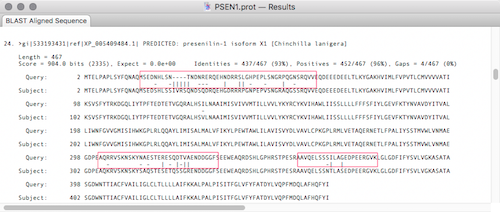
All these changes will be the new defaults until you reset them.
To change the line length
Remember that due to changes at the NCBI, BLAST and Entrez will only work in MacVector 15.1 and later.