MacVector 17 has a greatly improved Assembly Projects manager, for better organization of multiple sequencing datasets, multiple references sequences and repeated jobs. Every time you run a new assembly job (either a reference assembly or de novo). A new job object is created in the Assembly Project window contains resulting contigs and any unaligned reads from the assembly.
A new properties tab lists all of the relevant details for the currently selected job, including input parameters, total assembled reads, average contig length, total contig length and N50 to give you an idea of the quality of genome assemblies.
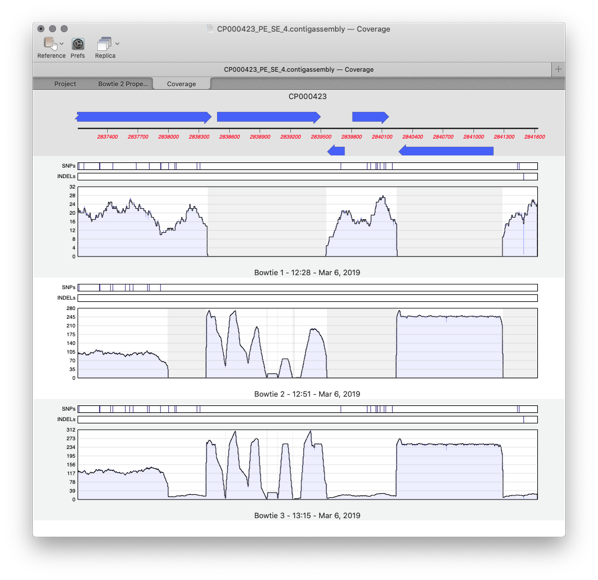
A new Coverage tab directly compare multiple reference assemblies. You can compare different sequencing datasets assembled against the same reference sequence with expression level comparison. Now that you can store multiple assembly jobs in a single Assembly Project, you can also directly compare multiple runs of the same dataset against a single reference sequence to compare which is the best assembly.
Simple DNA sequence assembly on a Mac with MacVector with Assembler.
MacVector has a software plugin called Assembler that integrates directly into the DNA sequence analysis toolkit and provides DNA sequence assembly functionality. Dealing with sequencing reads has never been easier.
MacVector includes no less than five different assemblers just a few mouse clicks away from your sequencing reads. Phrap assembles Sanger sequencing reads or existing contigs, while there are three separate NGS de novo assemblers – Velvet for short read datasets, Flye for Nanopore and PacBio long reads and SPAdes for mixed assemblies. For reference assembly Bowtie2 can map millions of sequencing reads against genomic reference sequences and is ideal for RNASeq gene expression analysis data too.
Assembler is tightly integrated into MacVector. It’s easy to bring sequencing reads into MacVector, and it’s just as easy to directly design primers for a contig, run BLAST searches on a contig, and much more, right from your desktop!