With MacVector Pro and Assembler you can use Bowtie to perform RNA-Seq analyses using NGS data. The interface even has specialized output tabs listing the coverage information and statistics for each annotated CDS and gene feature on the genome. You can download a short tutorial and a sample dataset that illustrate the analysis workflow using a small (1.6 Mbp) prokaryotic genome.
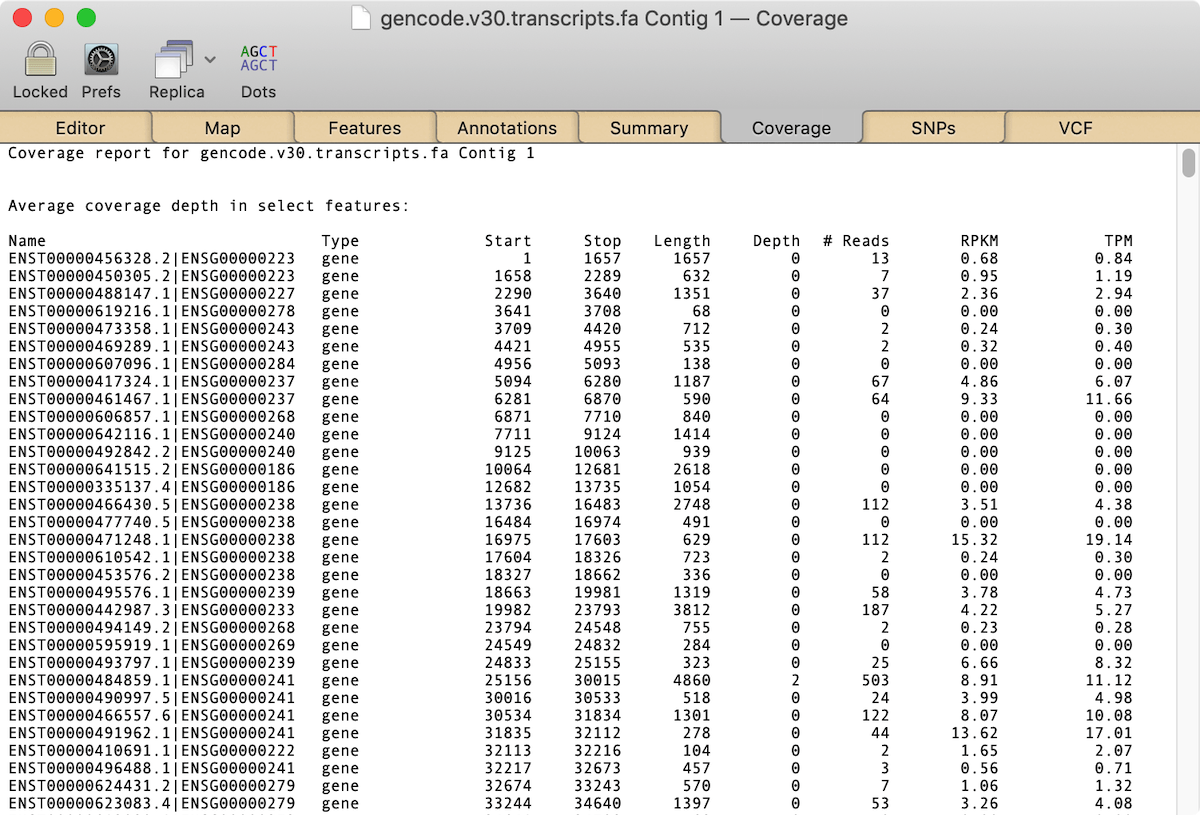
What surprises many people is that the combination of MacVector and modest Macintosh hardware can actually perform this analysis on the human genome. Now there are limitations to this – it’s not currently practical to do this with the entire genome due to memory and processing constraints, but it is possible to run an analysis against the known Human Transcriptome. The latest version of this can be downloaded from the GENCODE database. There is a new RNA-Seq Human Transcriptome Analysis Tutorial that describes the basic procedure in detail and some sample data that can be downloaded. The end result is that you get a table similar to that shown below that can be copied and pasted into Microsoft Excel for additional analysis.
Simple DNA sequence assembly on a Mac with MacVector with Assembler.
MacVector has a software plugin called Assembler that integrates directly into the DNA sequence analysis toolkit and provides DNA sequence assembly functionality. Dealing with sequencing reads has never been easier.
MacVector includes no less than five different assemblers just a few mouse clicks away from your sequencing reads. Phrap assembles Sanger sequencing reads or existing contigs, while there are three separate NGS de novo assemblers – Velvet for short read datasets, Flye for Nanopore and PacBio long reads and SPAdes for mixed assemblies. For reference assembly Bowtie2 can map millions of sequencing reads against genomic reference sequences and is ideal for RNASeq gene expression analysis data too.
Assembler is tightly integrated into MacVector. It’s easy to bring sequencing reads into MacVector, and it’s just as easy to directly design primers for a contig, run BLAST searches on a contig, and much more, right from your desktop!