Here’s an overview of the many different sequence assembly tools within MacVector. We also have tutorials to download.
To assemble various types of sequencing reads, follow these steps.
- Choose File | New | Assembly Project to create a new empty project file.
Then follow one of the following:
To create a de novo assembly from Sanger reads
- Click on the Add Reads tool bar button, then select the sequence files you wish to assemble and click on the Open button. Read(s) file(s) can also be drag and dropped on the open Assembly Project window.
- Click Phred to basecall sequences in the project. Note that if no sequences are selected, phred will be run on ALL of the files in the project.
- Click Phrap to assemble the reads.
To create a de novo assembly from NGS datasets
- Click on the Add Reads tool bar button, then select the sequence files you wish to assemble and click on the Open button. File(s) can also be drag and dropped on the open Assembly Project window. Paired reads files are automatically detected.
- Choose either SPAdes or Velvet to assemble the reads.
To create a de novo assembly from PacBio or Nanopore datasets
- Click on the Add Reads button to add PacBio or Oxford Nanopore reads in fasta, fastq or gzipped (.gz) format. File(s) can also be drag and dropped on the open Assembly Project window.
- Double-click on the Status column of the imported read file(s) and set the data type to “PacBio” or “Oxford Nanopore” as appropriate.
- Choose Flye to assemble all of the sequences of the project.
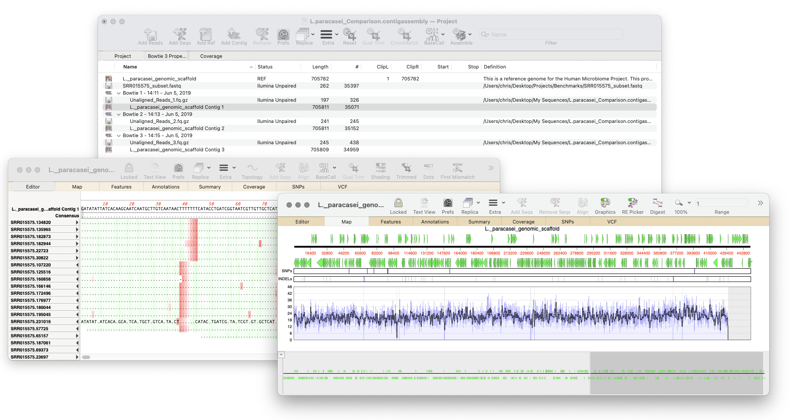
To create a reference assembly
- Click the Add Reads button, then select the sequence files you wish to assemble and click on the Open button.
- Click Add Ref, select the sequence file(s) you wish to align the reads against and click on the Open button.
- Click Bowtie or minimap to map all read files against all of the reference sequences in the project.
Not sure if you have Assembler? Choose MacVector | About MacVector. If the screen that appears says MacVector with Assembler, Pro Edition then you have it. If not, you can sign up for a fully functional 21 day trial version.
