If you have the Assembler module, MacVector can align millions of NGS reads from RNASeq experiments against large genomes and generate a coverage table displaying the relative expression levels of every gene in a genome. The key to this functionality is that you must have a reference genome with genes annotated as CDS or gene features – then you can use Bowtie to rapidly assemble reads from fastq files against the genome and generate a report listing the coverage for each feature. The steps to do this are:
- Use File | New | Assembly Project to create a new project.
- Click on the Add Ref toolbar button to add a suitable annotated reference genome.
- Click on the Add Reads toolbar item to add your RNASeq reads.
- Click on the Bowtie toolbar button to assemble the reads against the reference genome.
- Double-click on the resulting reference contig item and switch to the Coverage tab.
You will get a display similar to this.
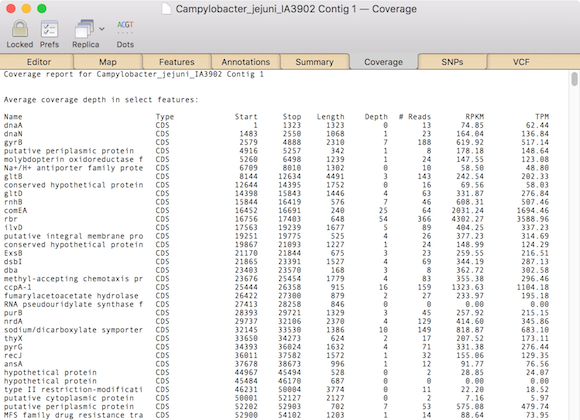
This lists each CDS and gene feature, along with the number of reads that aligned to each feature and the Reads Per Kilobase of transcript per Million mapped reads (RPKM) and Transcripts Per Kilobase Million (TPM) values that can be used to compare expression between genes and runs.
There is an RNASeq Expression Analysis Tutorial available that describes this functionality in more detail. If you are running MacVector 16.0.4 or later you will find this in the /Applications/MacVector/Documentation/ folder. Otherwise you can download the tutorial and sample dataset. Although this is a small dataset designed for rapid analysis, you can use this approach to align 50 million+ reads against the entire human transcriptome on a modest (16 GB RAM) MacBook Pro.
