Graphical Sequence Editing
Sequence Editing
Sequences can be edited using a fully functional Sequence Editor. As well as ensuring that feature locations are correctly maintained during edits, an audio function lets you playback selected residues for easy proofreading. Toolbar buttons let you view and edit the features, annotations and graphics associated with each sequence.
The sequence editor also acts as the primary window from which you can launch all MacVector analyses. All functions are integrated into a single MacVector application - no need to switch to a different module or program to find the algorithm you are looking for.
Graphical Features View
One of the strengths of MacVector is the ease with which high quality detailed graphical views of a sequence can be generated and copied into other compatible applications. MacVector uses OS X "Quartz" graphics and text for all of the Feature Graphics which can be copied into other applications like Photoshop and Illustrator with ultra-high vector graphic resolution.
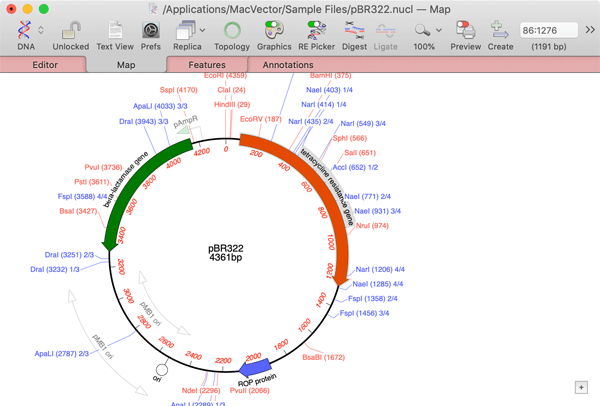
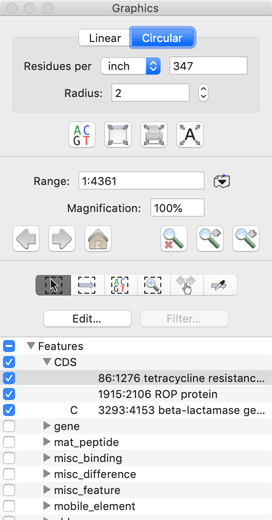
The visibility and general characteristics of the Map tab are controlled by a floating Graphiocs Palette and the displayed restriction enzymes are controlled by a highly interactive Restriction Enzyme Picker. Sequences can be viewed in linear or in circular format (great for plasmid constructs). When a feature item is selected, this also selects the corresponding region in the sequence editor and feature list view, greatly simplifying the selection of regions for copying or additional analyses.
Feature and Annotations Editing
Interactive lists of features and annotations can be viewed by clicking on the corresponding tab. MacVector considers features to be those annotations that have an actual location on the sequence. All other attributes (e.g. literature references or keywords) are considered to be an annotation. MacVector supports the full list of GenBank keywords and qualifiers. Creating a new feature is simple - just select the region of interest in the editor window, then click on the "+" button in the feature list window. Editing a feature is a breeze - double-click on a feature in the Map tab to open up the combined Feature/Symbol Editor; There's a very detailed tutorial that describes this in more detail.
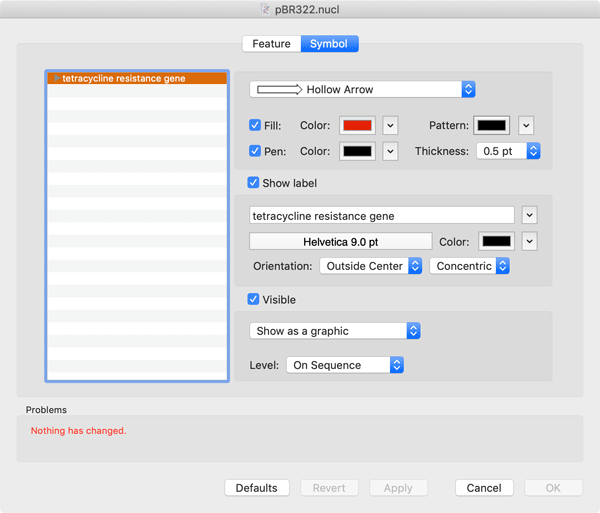
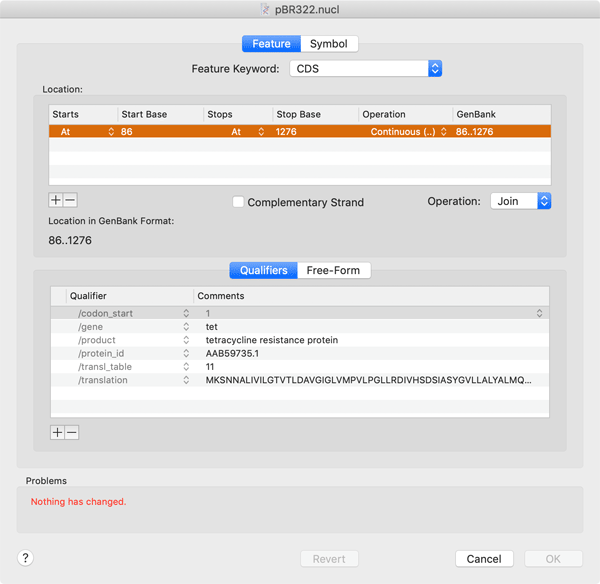
The Feature Editor tab is where you edit the GenBank settings of the feature. Here you can change the feature type or location and add appropriate quailifiers. MacVector is smart enough to guide you to only add those qualifiers that are relevant for the type of feature you are editing.
Click Cloning
Many of MacVector's analysis functions generate graphical result windows similar to the Map tab. In particular, you can use the Map tab and the Restriction Enzyme Search result window to replicate laboratory cloning experiments in silico. You can click on a restriction enzyme site, hold down the <shift> key and then click on a second site to select both sites along with the intervening DNA. When the selection is copied , the DNA sequence, enclosed features, graphical appearance information and the structure of the ends are all placed on the clipboard. You can then move to a second Map tab, select compatible target sites and paste in the copied DNA to create a new recombinant molecule. MacVector will "flip" copied molecules if required to match the target sites and will refuse to paste incompatible ends (although you can choose to override this if you wish). The main feature window also has a dynamic Restriction Enzyme Picker window that makes this even easier to accomplish. There are a couple of useful tutorials available that go into this in much more detail;
Clone Construction Tutorial
Gateway Cloning Tutorial
|

2x.png)


2x.png)
