 |
Interactive Restriction Enzyme Picker (RE Picker)
Introduced in MacVector 17, the RE Picker lets you dynamically filter the enzymes displayed in the Map tab of DNA sequence documents. By default, each time you open a DNA sequence, MacVector scans the sequence for ALL of the restriction sites in the selected restriction enzyme file, but only displays those that are selected in the file. However, all of the sites that meet the cut criteria are displayed in the RE Picker, and there are a number of sliders and controls that let you filter the enzymes displayed based on number of cuts, site size, cut site overhang type and selection status. This gives you very fine control over the enzymes that are shown and you can save sets of enzymes (e.g. all the non-cutters) for use in subsequent analyses.
The floating RE Picker is quite a large window in order to provide a reasonable visible list of enzymes, so there is a convenient button on the Map tab toolbar to show/hide the window so that it is only visible when you are interacting with it. A similar button controls the visibility of the floating Graphics Palette.
The RE Picker window consists of an upper area with a variety of controls and a lower area with a list of enzymes;
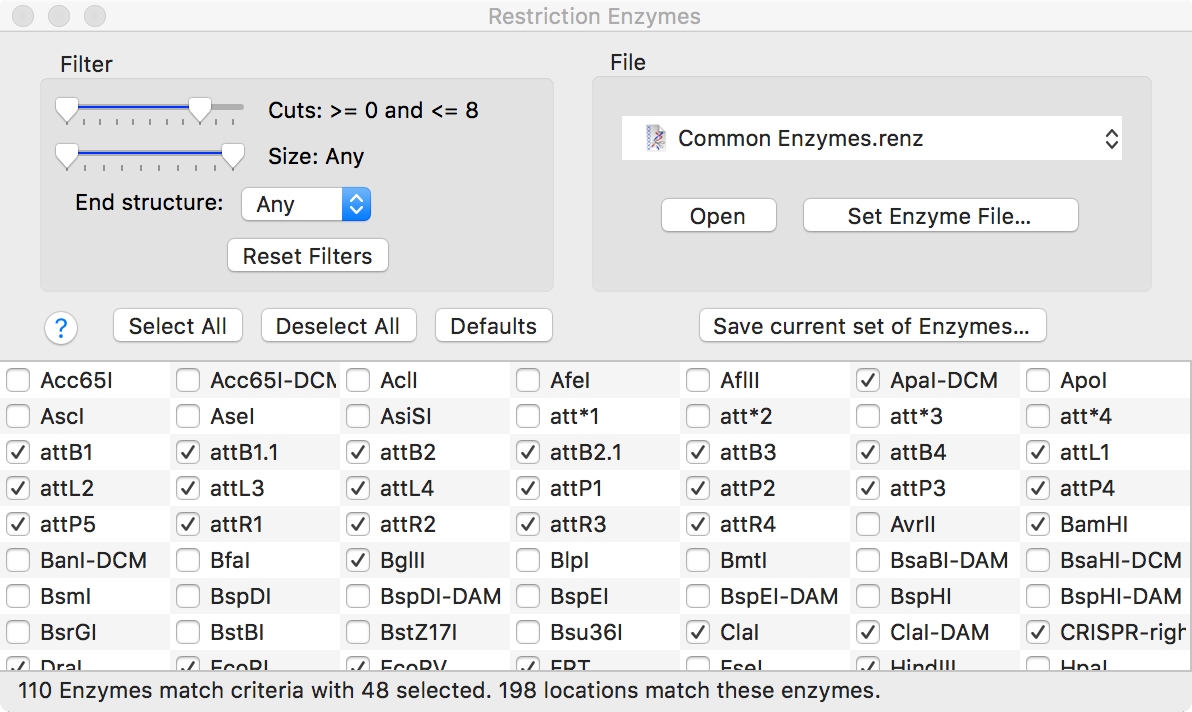
The Filter controls are dynamic, so that as you drag the sliders, the list of enzymes updates to reflect only those with the appropriate number of cuts, appropriate site size and end structure. At any time you can save the current set of enzymes to a separate .renz file. So, for example, you could adjust the sliders so that only enzymes with no cuts are shown in the list, then save those to a file, then use that file in a restriction enzyme analysis of a second sequence to identify enzymes that cut that, but that do not cut the first sequence.
|
 |

2x.png)


2x.png)
