I blogged about this a few years ago, but its something that still comes up on a regular basis. The blog link discusses the 6 main alignment algorithms in MacVector and how to decide which is the most appropriate for accomplishing different tasks. One common request we get is “I want to see my chromatograms/traces aligned so that I can edit the alignments”. For this, there are two main approaches. Note that you should NOT use the Multiple Sequence Alignment (ClustalW, Muscle or T-Coffee) interface unless you are truly looking for the evolutionary relationships between the sequences and know that they are already all in the correct orientation. None of these algorithms will “flip” the chromatograms when required, but the strategies described below will do that.
Typically, if you are aligning chromatograms it is because you are re-sequencing something. Admittedly, that is not always the case – if you are trying to determine the sequence of an unknown piece of DNA, then you need to use the add-on Assembler module. I’m not going to discuss that here but there is a tutorial that you can download from this link.
Otherwise, you would normally start with a Reference sequence – it might be the predicted sequence of the PCR fragment you cloned, the starting sequence for a mutagenesis experiment or even a related sequence from another organism. It can even be the entire genome of an organism to which you want to align chromatograms from sequencing runs of mRNA or cDNA clones. In any event, open that sequence first, then choose Analyze | Align To Reference. The window that opens contains a COPY of the starting sequence – now click on the Add Seqs button and select all the ABI/.ab1/SCF chromatogram files from your sequencing project directory to import them into the window;
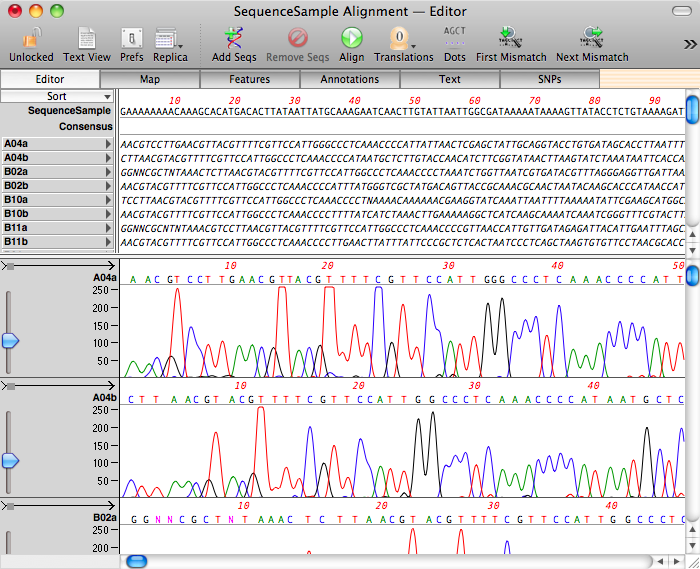
You can then click on the Align button to align the chromatogram sequences with the reference. There is a choice of two algorithms;
Choose cDNA Alignment only if you are expecting the presence of large gaps in the alignment. Typically, you would only use this algorithm for aligning cDNA sequences against a genomic sequence.
Use Sequence Confirmation when you expect the chromatograms to fairly closely match the reference sequence, with perhaps just a few small deletions/insertions due to sequencing errors or clonal variation.
Once you click OK, the algorithm will run and, crucially, chromatograms that align to the minus strand will automatically be flipped and aligned to the reference;
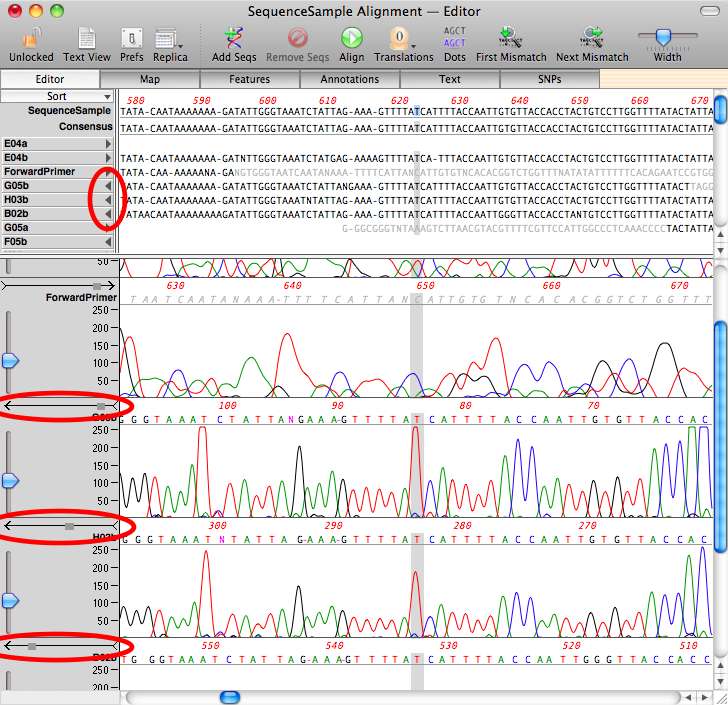
For more information on the use of Align To Reference, check out our tutorial that you can download from this link.
This is an article in a long running series of tips to help you get the most out of MacVector. If you want to get notified every time a new tip gets published, follow us @MacVector on twitter (or check the feed for the hashtag #101MacVectorTips) or like us on Facebook.