There are often times when you end up with a sequence containing gaps, especially if you make extensive use of the Align To Reference, Contig Assembly or Multiple Sequence Alignment interfaces to generate consensus sequences. You can select and copy the consensus sequence, or even individual aligned reads, from the Align To Reference and Contig editor tabs. The copied sequence can then be pasted into a sequence editor window, or a new document can be created by choosing File | New From Clipboard. In either case, if any gaps were present in the copied sequence, these will be preserved when pasted. MacVector does this because there are times when you might want those gaps preserved and, as you will see, it is a lot easier to remove them after the paste than it would be to type them back in by hand!
You can search for gaps and remove them using the Edit | Find | Find… menu item. The key to this functionality is that you must first check the Literal box before you can add a gap character in the Find box;
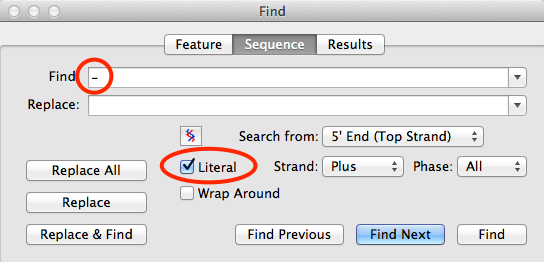
Just leave the Replace box empty, then when you click on the Replace All button, all of the gaps will be deleted. Note that if the Replace All button is inactive, that usually means that no gaps are actually present in your sequence.
This is an article in a long running series of tips to help you get the most out of MacVector. If you want to get notified every time a new tip gets published, follow us @MacVector on twitter (or check the feed for the hashtag #101MacVectorTips) or like us on Facebook.