MacVector 17 will be released later this week.
Get ready for our biggest release yet. MacVector 17 compares genomes, makes restriction enzyme cloning easier, automates the design of Gibson Assembly and Ligase Independent Cloning strategies. Makes plasmid maps even more beautiful and supports macOS Mojave’s Dark Mode to aid concentration on those late night primer design sessions!
For sequence assembly there’s a stack of new features. Align to Folder has a new Assembly Problems tab that identifies possible issues with your assembly (and what to do about them). For reference assemblies in Assembler there’s a new Coverage tab that allows you to compare multiple assemblies when mapped against the same reference. For example you can compare different sequencing datasets from different samples, the same dataset mapped with different parameters, or compare RNAseq datasets.
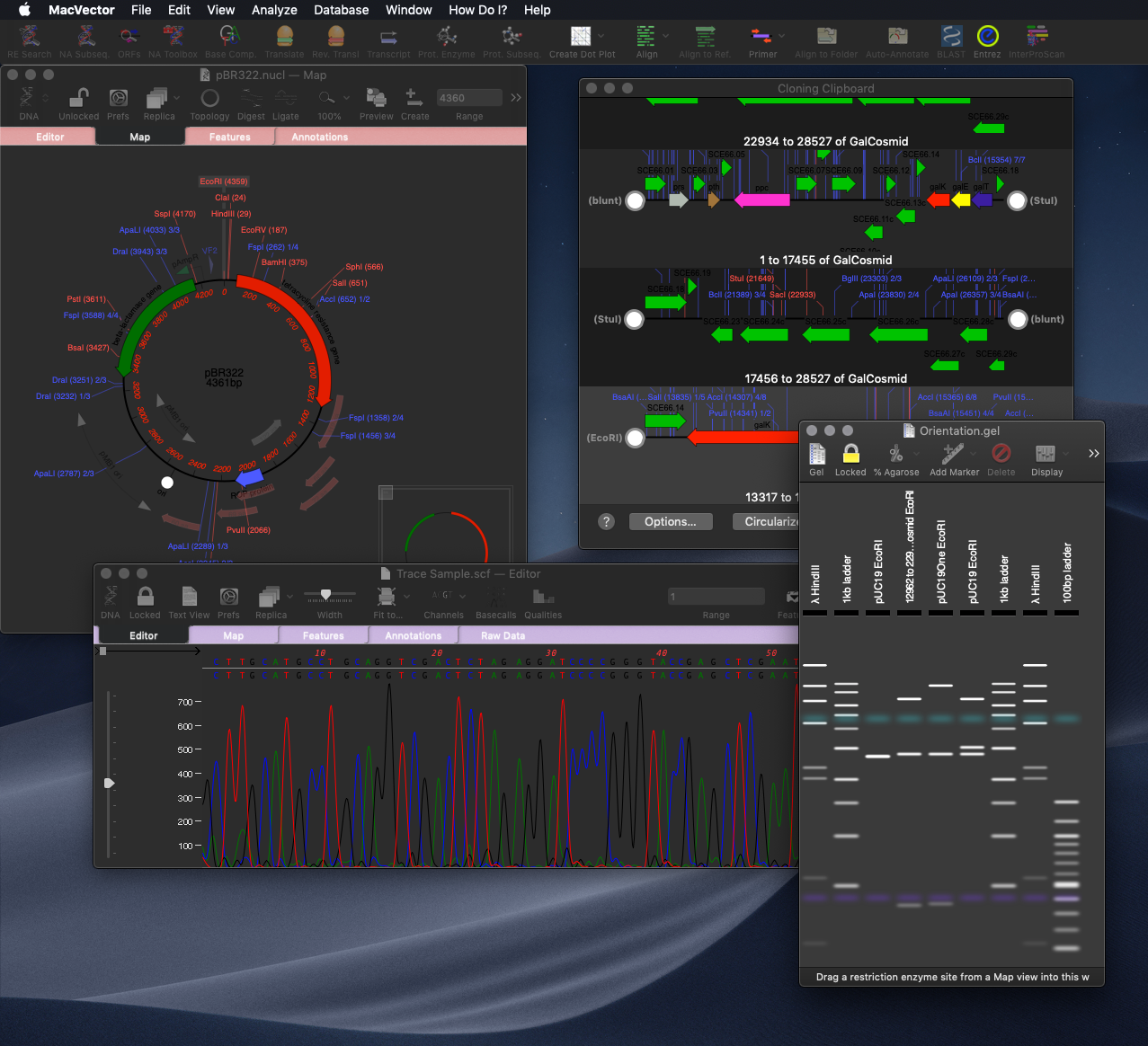
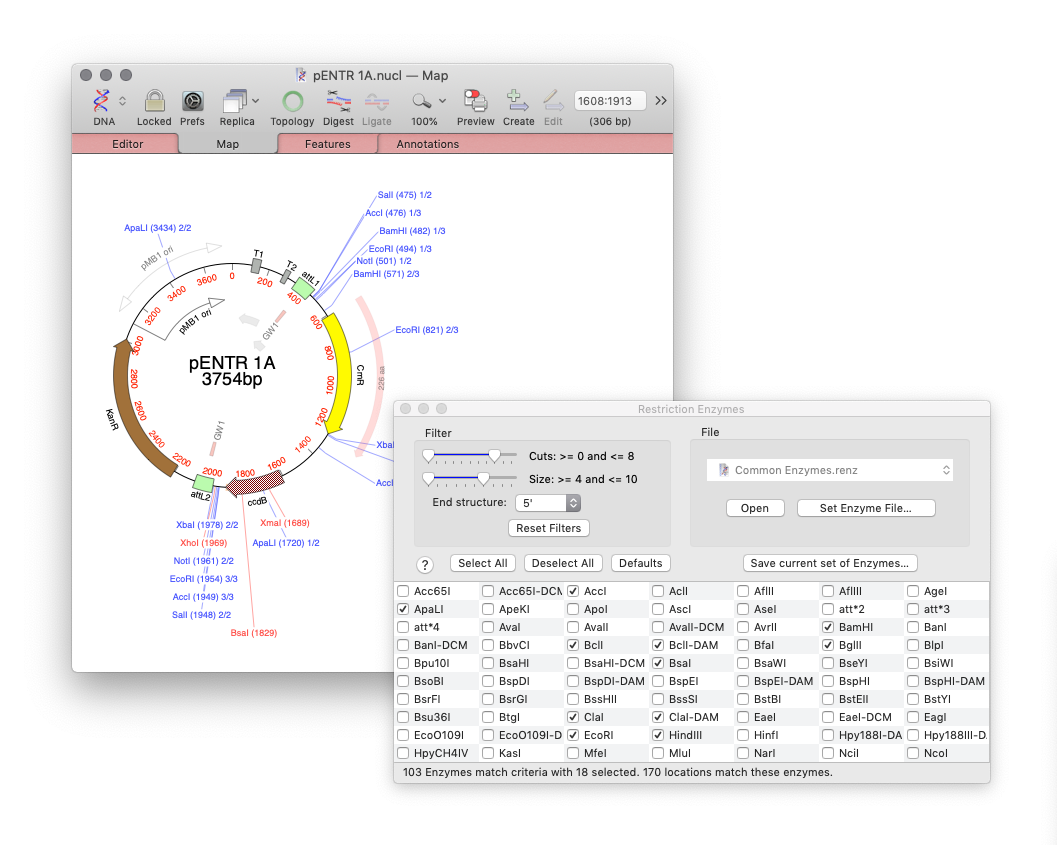
For cloning the new Restriction Enzyme Picker gives you an interactive way to quickly show what enzymes will digest your sequence. The new floating tool initially presents you with a list of all available sites in a sequence. However, you can filter on many attributes, such as number of cuts, 5’ or 3’ overhangs and blunt ends. What’s more is that you can take a set of cut sites from one sequence (for example an expression vector) as the input to digests of other sequences for easier optimizing of construct workflows.
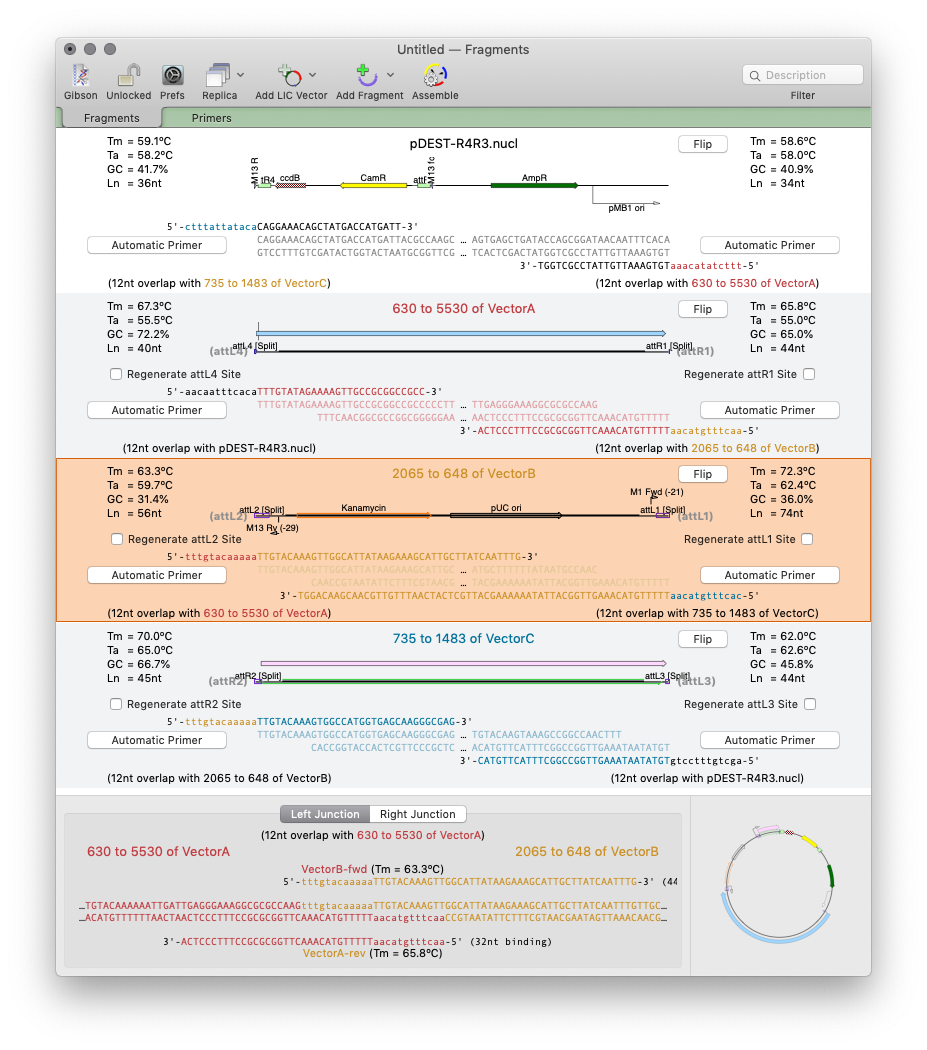
For non restriction enzyme based cloning the new Gibson Assembly/Ligase Independent Cloning tool provides automated design of ligase-independent cloning strategies. The tool supports 5’ exonuclease driven Gibson assembly as well as the T4 DNA Polymerase 3’ exonuclease “Ligase Independent Cloning” approach. It can automatically design primers when you specify fragments and vectors to use. Alternatively you can provide custom primers (manually or from tools such as the NEBuilder website). You can also just provide existing fragments with overlapping ends for MacVector to assemble. The interface shows the exact structure of the junctions between fragments, including relevant CDS translations for confirming the frame of protein fusions. The cloning strategy can be saved as a new cloning project document and finally automatically assembled into a single sequence.
Over the past few releases many new tools and features have been added to make fully annotated and beautiful plasmid map’s easier than ever. Continuing this theme MacVector 17 now utilises the power of the Primer Database and will automatically display your very own freezer drawer collection of oligos directly on your sequence, automatically! That means when you open even blank unannotated sequences you will see common features, RE recognition sites, putative ORFs and now your primer binding sites.
Scan for.. Missing Primers joins the existing Scan for tools that automatically display restriction sites, missing common features and putative open reading frames on your sequences.
This release is also aimed at making life easier for new users. There’s a brand new How Do I menu that shows common workflows with either simple step by step guides, or short videos. Plus every tool now has a link to a help topic on how to use it. MacVector has always been easy to use, but now even the most complicated tool has a simple way to get started… Quickly!
Finally we’re excited to announce that MacVector 17 supports macOS Mojave’s Dark Mode. When you are working late at night designing those constructs, then Dark Mode will help aid your concentration!
Here’s a full list of what’s new in MacVector 17
There’s also a more detailed list and the Release Notes.
Dark Mode Support: use macOS Mojave’s Dark Mode to aid concentration on those late night primer design sessions!
Scan For… Missing Primers: utilize the power of the Primer Database to display binding sites from your lab primer collection
Restriction Enzyme Picker: a new tool for easy identification of useful enzyme cut sites. Dynamically filter Restriction Enzymes to simplify the identification of useful enzyme cut sites. Use cut sites from one sequence as the input to others for planning construct workflows.
Compare Genomes: Compares two related annotated genomes to identify and list identical, similar and weakly similar features along with missing features.
Gibson/Ligase-Independent Assemblies: Automatic primer design and assembly for Gibson Cloning or Ligase Independent Cloning methods.
How Do I?: new menu shows common workflows with step by step guides. Every tool has a link to a video tutorial.
Assembly Problems: identify mis-assembled sequences based on excessive mismatches, discontinuous reads and other common problems.
Coverage Tab: Compare different datasets assembled against the same reference sequence with expression level comparison.
Redesigned Assembly Project: Easier organization of multiple datasets, multiple reference sequences and repeated jobs.
How to upgrade to MacVector 17.0
If you have active maintenance and are running MacVector 15.5.4 or later then you will be notified about the new release. To install this version, you must have a maintenance contract that was active on 1st November, 2018. You must also be running MacVector 15.5.4 and OS X 10.9 Mavericks or later.
If you have an older version of MacVector then download the trial and request an upgrade quote.
Even if you have downloaded the trial in the past then downloading a new trial will give you a fresh 21 days to evaluate MacVector.
When a trial license expires it becomes MacVector Free. So if you decide against upgrading then you can just delete the trial license and easily go back to your current version. It’s risk free as MacVector files are backwards compatible.