The Database | Align To Folder function is a very useful tool to find and retrieve similar sequences from folders on your computer or on other local machines. Think of it as your own personal BLAST service. It can not only search individual sequences in any format MacVector can read (MacVector, Genbank, EMBL, ABI etc) but will also process collections of sequences in fasta or fastq format.
One important factor to consider in these searches is the DNA Scoring Matrix (.nmat) file to use. There are several included in the /Applications/MacVector/Scoring Matrices/ folder. The default file is DNA database matrix.nmat. This is ideal for identifying sequences that are not particularly closely related, such as the same gene from distant organisms or sequences matching a highly degenerate input sequence such as a reverse translation of a protein sequence.
However, one common use of Align To Folder is to identify and retrieve NGS reads from large collections in fasta or fastq formatted files. It is particularly useful for finding reads to help resolve repeat regions or close gaps between contigs. When running these types of alignments, it is preferable to use a different matrix that is more tuned to finding reads with a greater identity stringency. The best scoring matrix for this is DNA identity with penalties matrix.nmat. Here’s some examples, using a short query sequence, where the searches differ only in the scoring matrix.
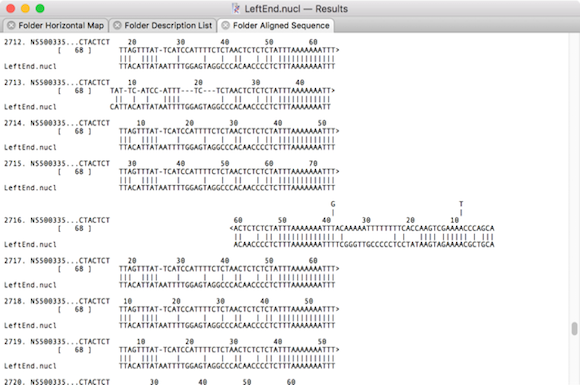
Low-scoring alignments using DNA database matrix.nmat
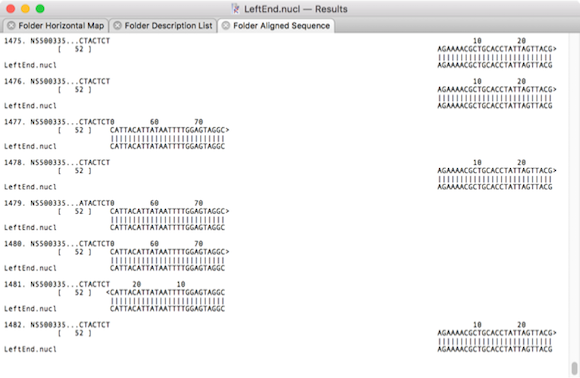
Low-scoring alignments using DNA identity with penalties matrix.nmat
It can clearly be seen that the second example has true matching alignments that represent sections of reads that extend beyond the query fragment. All of the reads can safely be retrieved and used in additional assembly analyses to extend the query or help resolve repeats. However, the DNA database matrix example contains matches that have extensive regions with very poor similarity. These clearly do not represent reads that could be used to extend the sequence of the query sequence.
For additional information about possible uses of Align to Folder, check out this blog post.