The CDC recently published diagnostic real-time primers for identification of SARS-CoV–2 in any person suspected of having COVID–19.
Unfortunately as pointed out on the Biome Informatics blog these primers have issues that should have easily been detected had the primers been tested using a good quality primer testing tool (the linked blog post uses Primer3). What’s more is that if a good quality primer design application, such as MacVector’s Primer3 tool, had been used from the beginning, then these issues would never have occurred.
Some primer design software tools can be difficult to use. However, MacVector provides an easy to use interface to Primer3. Designing a pair of primers to amplify a target can be as simple as just three mouse clicks (click on the target gene, click ANALYZE | PRIMER3 and then click OK).
MacVector also has QuickTest primer for visual design and testing of primers. QuickTest Primer simplifies primer design by showing your primer and its statistics in realtime. Does your primer have a hairpin? Nudge it along your template until the hairpin goes? Want to add a restriction site? Then add one and again nudge your primer to optimise the oligo.
MacVector also has built in tools for helping find suitable targets to amplify using Blast or scanning local sequences too.
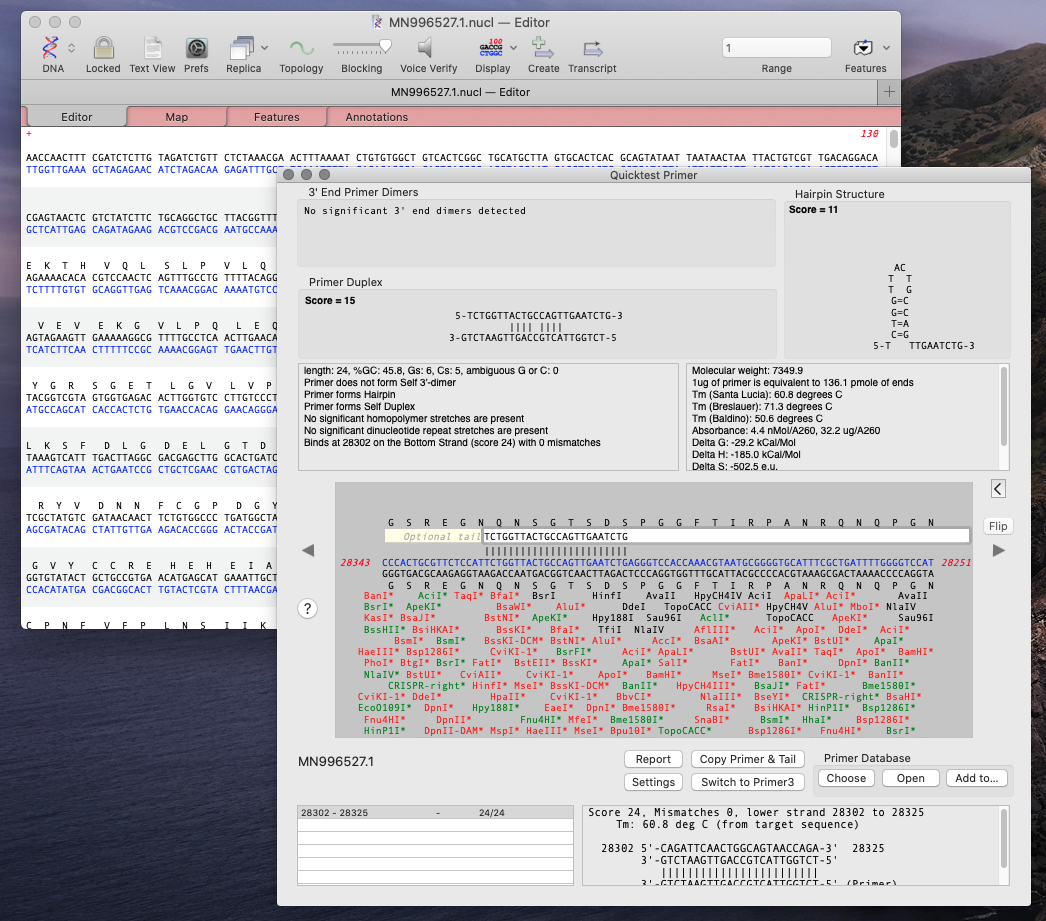
Using MacVector’s Quicktest Primer tool you can see the obvious hairpin in the reverse primer of the CDC first set of primers.
Here’s a demonstration workflow on how to use MacVector’s Primer Database and Quicktest Primer tools to look at these primers:
Download the primers
- Download the CDC primer list (in a MacVector Primer Database file).
- Open MACVECTOR | PREFERENCES | SCAN DNA | PRIMERS.
- Click SET DATABASE FILE and choose the downloaded file.
Download one of the many sequenced SARS-CoV-2 genomes
- Open MACVECTOR | ENTREZ and search for “organism=SARS-CoV-2” and “all fields = genome”.
There are a lot of submitted genomes from this organism (94 as of March 16, 2020).
- Double click on a hit to open the genome directly in MacVector.
- Now switch to the MAP tab.
You will see the primers annotated on the sequence (faded as they are dynamically shown).
- Open ANALYZE | QUICKTEST PRIMER
- Click the Insert from Primer Database button (as seen in the screenshot below).

You will see the primer displayed in the QT Primer interface showing any flaws. Most of the primers have hairpins.

Designing Primers
Here’s an example workflow of how you would design primers to detect the N gene.
Please note that for this demonstration workflow we have designed primers against the N gene, that had been selected by the CDC as a target. For real world purposes you would need to select an appropriate target. One way would be to align all sequenced human CoV-2 genomes (or whatever organism you are looking for), then look for highly conserved regions. Particularly in regions of the virus thought to be critical for pathogenicity. Those conserved regions can then be used as targets to design primers. It would also be useful to also select regions that would distinguish SARS-CoV-2 from the original SARS-CoV-1.
- Select the N Gene in the SARS-CoV-2 genome (the one you downloaded above).
- Run ANALYZE | PRIMER DESIGN/TEST (Primer3)
- Change the setting to REGION TO SCAN and enter a product size of 400 to 500
- Check Hybridization Probe Sequence.
- Ensure all primers are set to Find Primer and not USE THIS PRIMER.
- For this workflow example you do not need to change any advanced options.
- Click OK
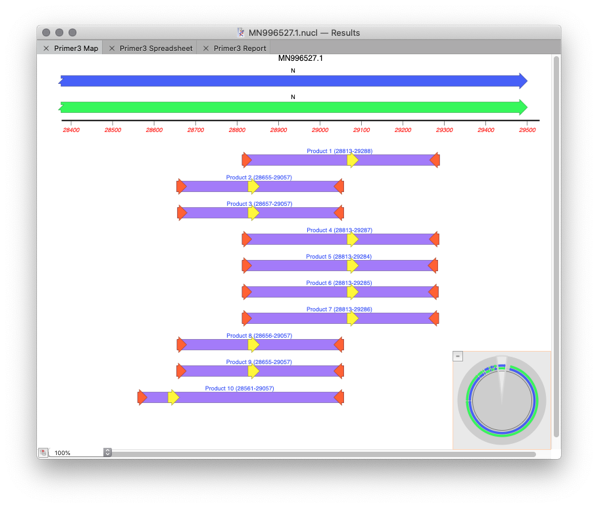
In the Results window you will see a MAP with the generated primer pairs and products.
The initial left primer still has a hairpin. You could tweak Primer3 settings. However, you can quickly tweak the primer itself using Quicktest Primer. If you slide the primer three bases to the left then the hairpin will go.
- Select the LEFT primer in the Results Spreadsheet tab and choose ADD TO PRIMER DATABASE
- Choose a suitable name.
- Repeat for the RIGHT primer and the PROBE.
- Open QuickTest Primer
- Click INSERT FROM PRIMER DATABASE and test each primer.
- To slide a primer left or right click on the cursor buttons either side of the primer.
- Don’t forget to save any modified primer. You may also want to test the primers again using primer3. If you save the primers to your Primer Database then this is easy to do.
