Our upcoming release, MacVector 12.6, has just entered beta testing. As usual we’ve added a mixture of new features as well as improvements to existing tools. One tool that we are excited about is a realtime primer design tool. This allows you to manipulate your primer and see secondary structure and other useful oligo statistics as you edit it.
Primer Design Enhancements
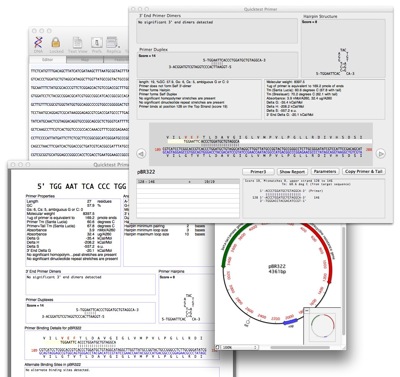
MacVector 12.6 introduces a new “Quicktest Primer” interface that simplifies the design of primers with mismatches and/or tails and complements the existing primer and probe design tools. This can be used in conjunction with the existing Primer3 tool to design pairs of primers with mismatches and/or tails and to easily generate the predicted product of the reaction, complete with tails and/or mismatches.
Other useful features of QuickTest Primer are:
- View secondary structure in real time as you enter a primer sequence
- Generate a detailed report of your primer
- Slide your oligo along your sequence to find the optimal site.
- See any changes in the translated protein sequence as you design your primer
All primer/probe design tools now share the same reaction conditions. So regardless of which tool you use you only have to set these once.
Importing data into MacVector
Making it easy to bring data from public databases into MacVector is always one of our priorities. Adding the ability to import annotation from the UCSC’s Genome Browser and similar sites such as Wormbase and ToxoDB has been a popular user request over the past few years. All these sites allow the export of data in one of the annotation formats such as General Feature Format (GFF) or BED and now MacVector will allow you to import the features in these files and annotate an existing sequence.
This allows you to continually update your own working copies of your favourite sequences with publicly available data.
- Support of GFF, GFF3, and BED files to annotate blank sequences.
- Create new sequence(s) with complete annotations by copy and paste of GenBank, EMBL and FastA documents.
Assembler improvements

Following the introduction of NGS reference assembly using Bowtie in MacVector 12.5 there have been further changes in response to user feedback. There has also been major performance enhancements to the display of large assemblies.
- Import BAM/SAM alignments for display.
- Importing ACE files for displaying alignments created by other assemblers.
- de novo assembly has been optimised further for size and speed.
- Better support for undoing assemblies.
- Child contigs are annotated as features on the reference contig.
- Interactivity between contigs so that selections in a child contig are reflected in the reference contig.
- Reported SNPs are annotated to the graphical Map View.
- Child Contigs are also listed in a text report.
Performance enhancements

- Pustell Matrix has been enhanced for genome vs genome comparisons. Now you can do a genome vs genome comparison of two E.coli genomes in less than 5 mins on a recent Mac.
- Major performance enhancements to Map Graphics for easier viewing of annotation rich genome sequences.
