You can import BAM files, containing reads mapped against a reference sequence, into a MacVector Assembly Project. As well as the BAM file(s) you will also need the original reference sequence the reads were mapped against. FASTA is fine, but an annotated reference is better for visualisation.
The tool needed is called ADD CONTIG. This is one of the toolbar buttons in an Assembly Project:
First create a new assembly project.
FILE > NEW > ASSEMBLY PROJECT
click ADD REF to add the reference sequence.
Use ADD CONTIG to import your BAM/SAM file.
Then you need to associate the BAM file(s) with the reference:
– select the reference and an imported contig(BAM file).
- Right click on and select UNITE REFERENCE WITH CONSENSUS SEQUENCE
You can optionally also generate a report on any variants (either at the previous step or a later stage).
- Right Click and choose GENERATE VCF
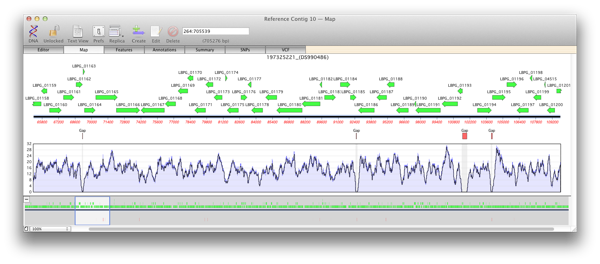
If you import multiple BAM files against the same reference sequence you can also graphically compare these datasets with the Coverage Tab (third tab along in the Assembly Project window).

Incidentally if you need to access the BAM files from within MacVector’s Assembly Projects then you can right click on an Assembly Project and view the contents.
Simple DNA sequence assembly on a Mac with MacVector with Assembler.
MacVector has a software plugin called Assembler that integrates directly into the DNA sequence analysis toolkit and provides DNA sequence assembly functionality. Dealing with sequencing reads has never been easier.
MacVector includes no less than five different assemblers just a few mouse clicks away from your sequencing reads. Phrap assembles Sanger sequencing reads or existing contigs, while there are three separate NGS de novo assemblers – Velvet for short read datasets, Flye for Nanopore and PacBio long reads and SPAdes for mixed assemblies. For reference assembly Bowtie2 can map millions of sequencing reads against genomic reference sequences and is ideal for RNASeq gene expression analysis data too.
Assembler is tightly integrated into MacVector. It’s easy to bring sequencing reads into MacVector, and it’s just as easy to directly design primers for a contig, run BLAST searches on a contig, and much more, right from your desktop!
