In our latest release, MacVector 18.5, we added a new tool to call heterozygotes in sequencing reads.
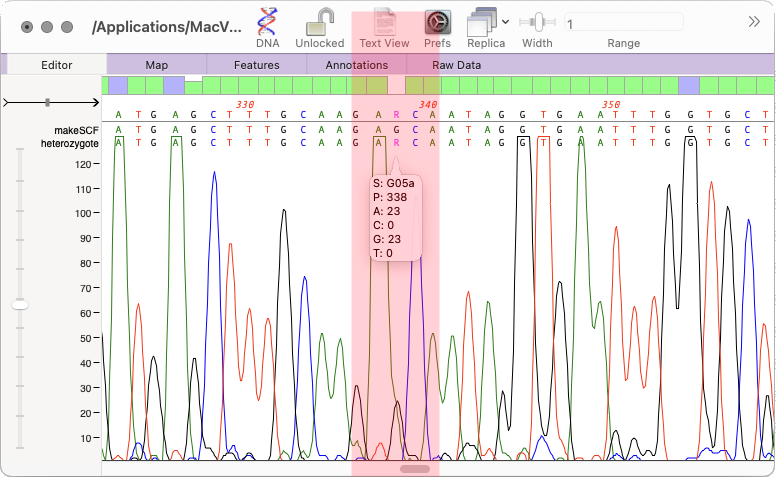
The heterozygote analysis tool allows you to either view heterozygotes in Sanger trace files or to permanently change the basecalled sequence with an ambiguity representing the called heterozygote. The tool works on multiple trace files in the Assembly project manager or the Align to Reference editor. You can also run it on a single trace file in the Single Trace Editor
How to run Heterozygote Analysis
To view putative heterozygotes
Select Trace files in the Align to Reference editor or Assembly Project Editor or open a trace file in the the single trace sequence editor.
- Run ANALYZE – HETEROZYGOTE ANALYSIS
- An Options dialog will appear. Change the options and click OK
- A summary dialog will appear showing the number of heterozygotes found across how many sequences.
- Click OK
- A new tab will appear in the Results window showing the location of the possible heterozygotes.
You can click on the highlighted blue position to be taken to the heterozygote. Note if you are in Assembly Projects or Align to Reference editors then the heterozygote will be displayed in the Single Trace Editor.
To permanently basecall the sequence
Assembler
- Add trace files to your Assembly Project
- Select one more more trace files
- Select ANALYZE | BASECALL | USING HETEROZYGOTE ANALYSIS
- An Options dialog will appear. Change the options and click OK
- A summary dialog will appear showing the number of heterozygotes found across how many sequences
- Click OK
Align to Reference
- go to FILE | NEW | ALIGN SEQUENCES TO A REFERENCE
- Choose your reference sequence
- Add trace files to your Assembly Project
- Select one more more trace files
- Select the BASECALL toolbar button or ANALYZE | BASECALL | USING HETEROZYGOTE ANALYSIS
- An Options dialog will appear. Change the options and click OK
- A summary dialog will appear showing the number of heterozygotes found across how many sequences
- Click OK
Trace File Editor
- Double click to open a trace file.
- Select ANALYZE | BASECALL | USING HETEROZYGOTE ANALYSIS
- An Options dialog will appear. Change the options and click OK
- A summary dialog will appear showing the number of heterozygotes found.
- Click OK
Once run there is a new BASECALL line showing the new sequence. In the Assembler Project editor an H will appear in the status column. Heterozygotes are indicated by an ambiguity.
Ensure the BASECALL toolbar button is toggled to green to view basecalled lines.
Settings
The default settings should be enough for the majority of trace files. However, there are a number of settings that can be adjusted where needed. The default value is in brackets.
- Normalise peak heights using a window of (25) residues
- Percent of each peak width to use (50%)
- Minimum number of normalised residues (3)
- Minimum Heterozygote threshold (35%)
- Minimum Base Call threshold (75%)
- Ignore low quality regions (yes)
- Where a window of (21) residues
- Has an average quality value of less than (20)

