Assembler is fully integrated into MacVector and allows you to manage sequencing data with the familiar MacVector ease.
de novo sequence assembly using Phrap, Velvet and SPAdes with Flye for PacBio and Oxford Nanopore.
Reference Sequence Assembly: Map millions of reads against genomes, transcriptomes or other reference sequences using Bowtie2.
Compare Genomes: Compare two related annotated genomes to see common or missing genes.
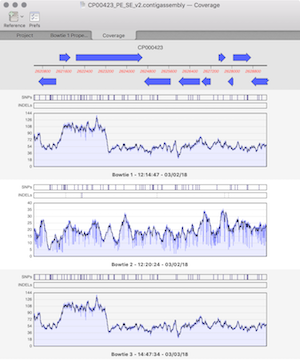
Coverage Tab: compare multiple assemblies with expression level comparison.
Variant Calling: SNPs and INDELS are visualised on your assembly and supplied in VCF.
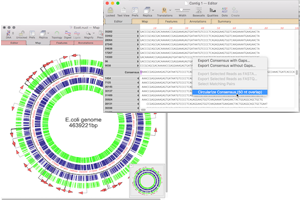
Bacterial genome tools Tools for finishing bacterial genomes including circularizing genomes.
Easy to use interface. Navigating around your assemblies has never been easier. Display an entire contig in the graphical Map and select a read to zoom straight to that region. Click on a base in a contig and see coverage and variants.
Heterozygote analysis. Analyse heterozygotes in Sanger trace files and assemblies.
Assembly Project manager makes it easy to assemble multiple datasets, reference sequences and assemblies. Work with unaligned reads from an assembly directly in the project.
Compare different assemblies. Map multiple datasets against the same reference.
RNA-Seq analysis with read depth visualization and per gene coverage data (RPKM & TPM).
