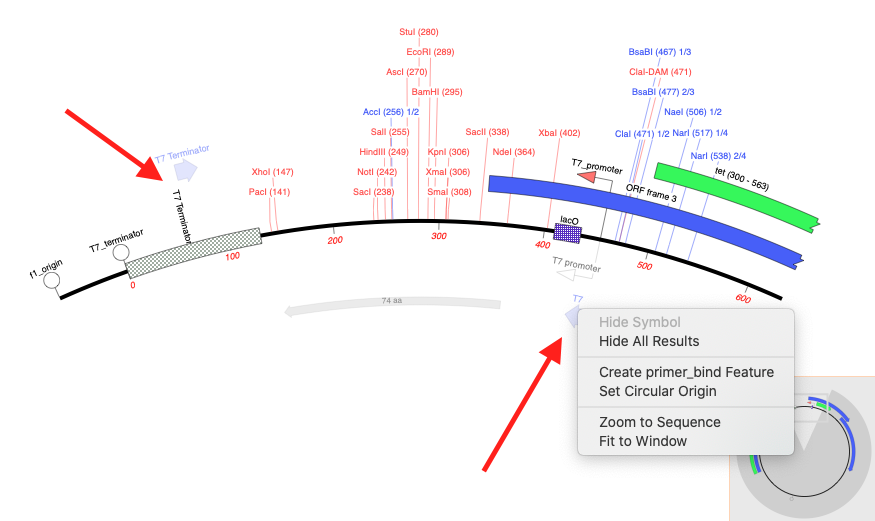
One new feature in our MacVector 17 release is the ability to automatically display primer binding sites in each DNA sequence that you open. Here’s an example of a couple of primers displayed on the popular pET 47b LIC cloning vector on each side of the LIC cloning site. The image shows how MacVector 17 has also been optimized for macOS Mojave “Dark Mode” and also includes a right-click popup menu that lets you quickly annotate interesting binding sites with a simple mouse click.
The feature is controlled from the MacVector | Preferences | Scan DNA | Primers tab. My default, it uses the Primer Database.nsub file that you can find in the /Applications/MacVector/Subsequences/ folder, but you can point it to any .nsub file of your choosing.
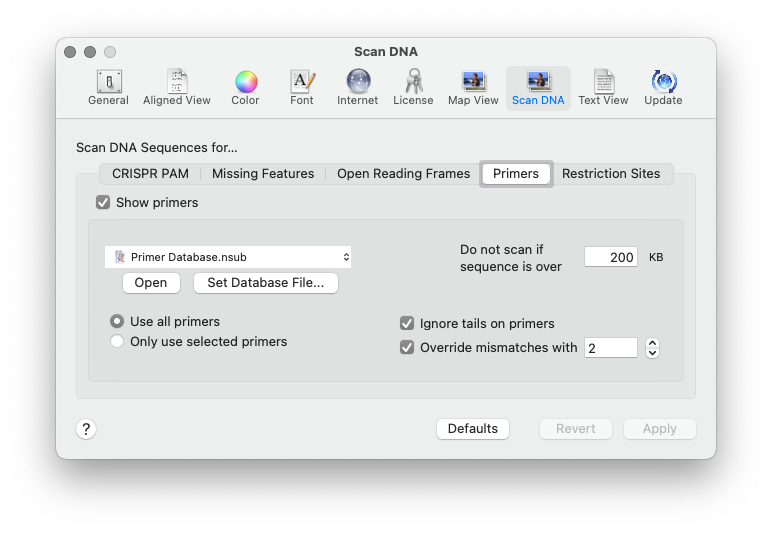
Scan for.. Missing Primers joins the existing Scan for tools that automatically display restriction sites, missing common features and putative open reading frames on your sequences.
