Our next release, MacVector 11, will be released this week. Continuing our policy of continuous improvement there’s a lot of under the hood improvements to make MacVector both faster and easier to use. However, MacVector 11 has a lot of other changes and some really useful new features that make this one of our best releases yet!
Here’s a short overview of these:
Auto Annotation.
How often have you received or downloaded a vector or other DNA sequence that has no annotations? The new auto annotation function in MacVector 11 will scan a folder full of existing annotated sequences and automatically add matching features to the bare sequence. Not only does the algorithm add the features, but it also copies the appearance information of the matching features so you can be assured that e.g. an ampicillin resistance gene is always a blue arrow, an M13 origin is always a striped red box etc.
Click Cloning Enhancements
In MacVector 11 you can manipulate the ends of restriction fragments before joining them together. A new interface lets you cut back or fill in each end of the source or target molecule before ligation.

Floating Analysis Toolbar.
For those users who prefer to click on toolbar buttons to initiate analyses, MacVector 11 introduces a new floating toolbar window containing buttons for all common MacVector analysis functions. You can customize the toolbar to show just the functions you use most often or show them all for rapid access to every available algorithm. There’s also a bunch of new toolbar buttons to add to sequence windows. These will allow you to put your own most commonly used buttons specific to both nucleic acid and amino acid windows. Giving you easier access to functions.
Sequence Editor Changes
The primary sequence editor has been rewritten using modern OS X code to better handle long sequences and to provide a more modern look and feel. In addition, you can now display 3 or 6 frame translations below the sequence.
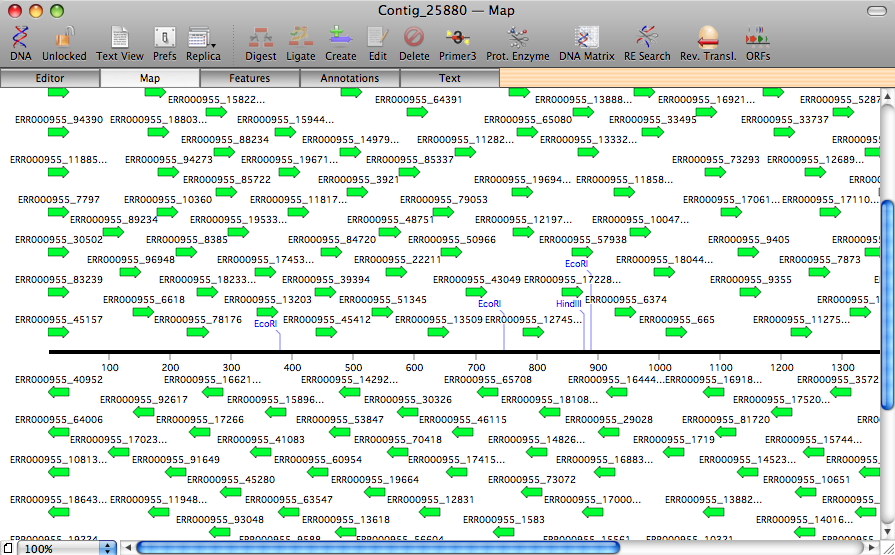
Next Generation Sequencing
The optional add-on Assembler module has been enhanced to provide support for next generation sequencing machines. Short read data may be imported in Fastq format and assembled using the latest version of phrap.
Miscellaneous Enhancements.
As always, we add a slew of minor enhancements to each release of MacVector designed to improve workflows, speed up processing or provide better integration with the operating system. Look for the ability to change the colors of chromatogram file displays for red/green color blind users, improved import of sequences from Vector NTI and better handling of genomic sequences.
Incidentally if you are still using an old dongle controlled version of MacVector you may be interested in an offer we have to upgrade!
