You can use the Database | Align To Folder function to scan large fasta or fastq files containing NGS data to find and retrieve just those reads that match a specific target sequence. The search is aware of paired-end reads, so when you retrieve hits, both reads of a pair will be saved into a pair of fasta or fastq files, even if only one of them matched the query sequence. This is a great way of finding sequencing reads to extend any short sequence. For optimum performance, set up the Align To Folder search like this.
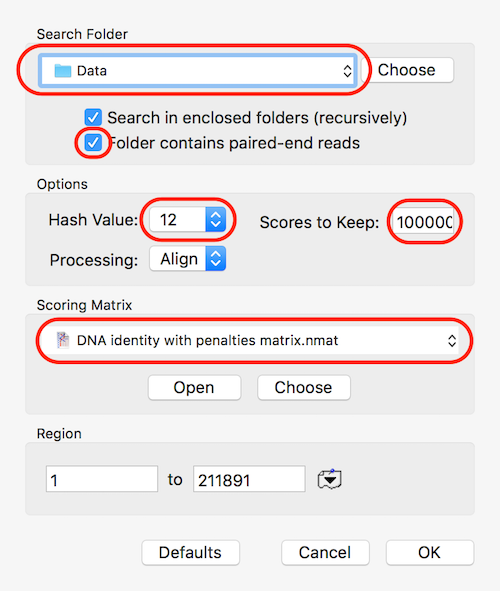
Set the Search Folder to the location of your data and select the Folder contains paired-end reads checkbox if you are working with paired data. For speed, make sure Hash Value is set to the maximum (currently 12) and use a large Scores to Keep value to make sure you can retrieve all the hits. Finally, use the DNA identity with penalties matrix to optimize the search so that only very close matches are reported.
On a moderate machine (e.g. a three year old 2.7 GHz i7 MacBook Pro), a search of 20 million x 90nt reads with a 500 bp search sequence might take from 45 mins to two hours, depending on the number of hits encountered. To retrieve the hits, select the numbered rows in the Folder Description List results tab and choose the Database | Retrieve to File… menu item. If you used paired-end data, two files will be produced, –1.fastq and –2.fastq, that you can then use as input to Assembler for SPAdes or Velvet assembly, or to Analyze | Align To Reference for more detailed reference alignment analysis.
