You can align hundreds, or even thousands of protein sequences within MacVector using three different alignment algorithms – ClustalW, MUSCLE or T-Coffee. Once aligned, you may be familiar with the colorful display in the Editor tab.

But there’s more to this than pretty colors. The default Color Group in MacVector is one called “Chemical Type”. In this, glycine, leucine, isoleucine, valine and alanine are all considered to be of the same type, and thus are included in the same group. You can access the Color Group selector/editor by clicking on the Groups toolbar button (note you may need to resize the window larger to see this button). The selector is also accessible from the Prefs | Consensus pane.
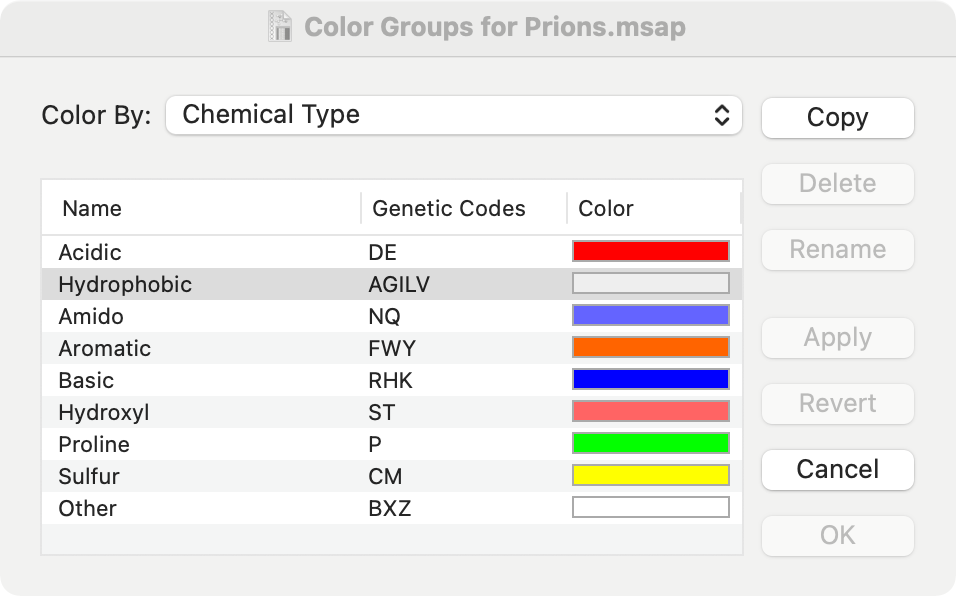
You can change the selected Color Group from about 20 built-in groups using the Color By: dropdown menu. You can edit the groups or even create your own groups. When amino acids belong to the same group, MacVector considers them to be “similar”. This affects many related functions throughout the multiple alignment interface. One example in the first image is a consensus that is a dot – none of the residues individually exceed 51% (the default identity threshold), but all belong to the same Color Group. If you select a different Color Group scheme, not only will the Editor tab update with the new colors, but the consensus will change to reflect the new groups.
The Picture tab also handles similarities – you can shade and outline residues based on the currently selected color grouping scheme. This is controlled from the Prefs | Picture Shading tab.
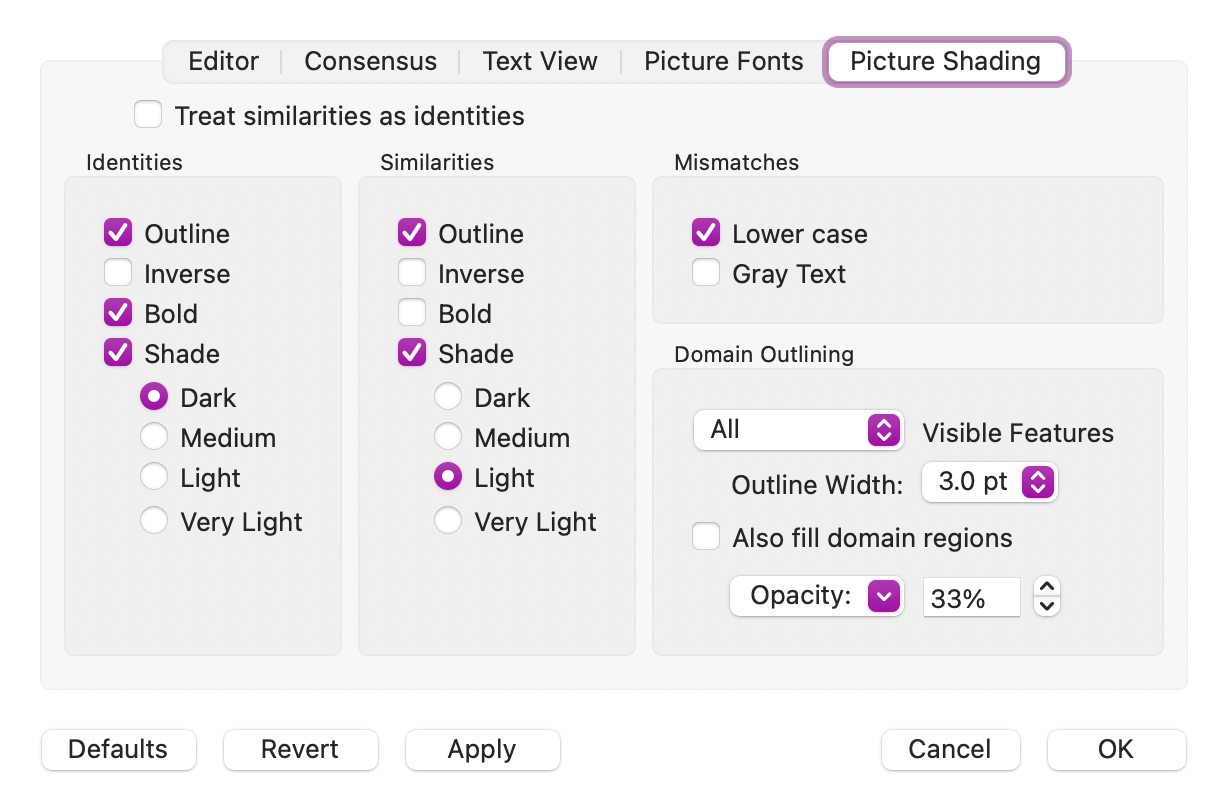
Finally, the Text, Pairwise and Matrix tabs also respond to the currently selected Color Group to determine similarities. Prior to MacVector 18.2.5, these always used the ClustalW Default Groups similarity scheme, but now, for consistency, they honor the currently selected group.

