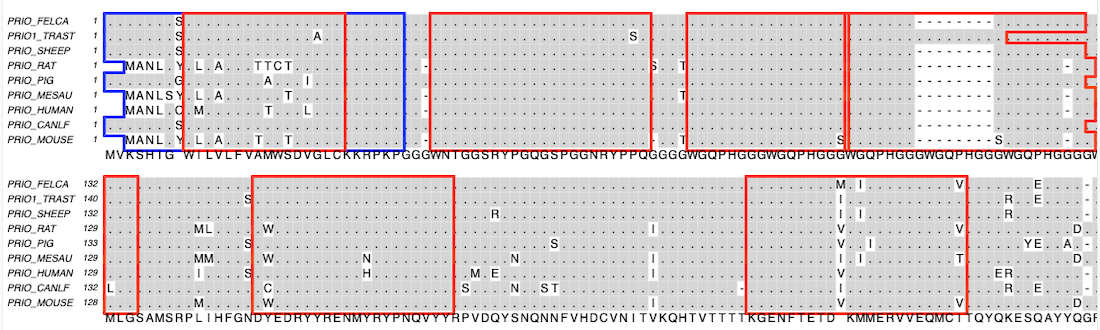
MacVector has a domain-outlining facility for multiple sequence alignments, letting you easily visualize the relationships between features in aligned protein sequences.
MacVector’s new multiple alignment file format retains the features/annotations from the sequences that are used to create the alignment. The colors of features from the individual sequence documents are used to outline the domains in the alignment. You can also create new domains and dynamically show/hide features in alignments.
Note that alignments created using versions of MacVector before 17.5 will not have this information and will need to be recreated.
Displaying shared domains:
- Ensure your protein sequences are annotated -Tip: you can use DATABASE | INTERPROSCAN to quickly scan and annotate domains to your proteins.
- Ensure the domains/features you are interested in are visible and set the Fill color to the color you would like to see in the alignment.
- You can also control the visibility of domains/features using the floating feature palette seen in the EDITOR tab.
- Use FILE | OPEN and select multiple protein sequences.
- Click OPTIONS (bottom left hand corner) and choose OPEN MULTIPLE SEQUENCE FILE – AS MULTIPLE ALIGNMENT
- Click OPEN
- Now run the alignment by clicking ALIGN
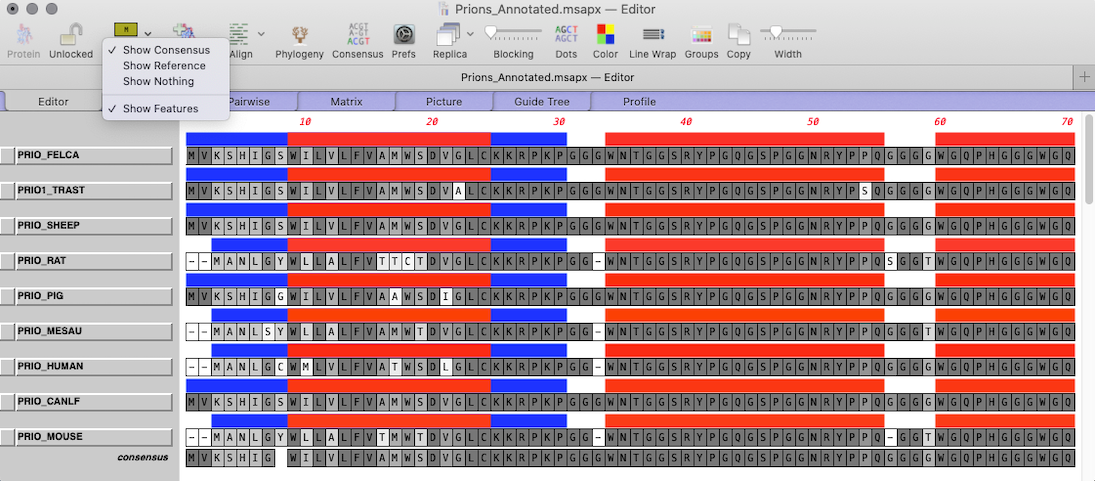
- In the EDITOR tab using the toolbar button turn on the feature display MODE to SHOW FEATURES
In the Editor tab a new line will appear above each sequence displaying the extent and color of visible features.
When you switch to the Picture tab, you will see colored outlines around the shared domains.