MacVector has a Agarose Gel interface which allows you to view photo-realistic recreations of restriction digests of linear and circular DNA molecules. The gels look so realistic that users have had a hard time telling photos of their own digests from the simulation in MacVector. When you first use the new tool and compare it to your real gels, you’ll see what they mean!
The Agarose Gel tool uses published algorithms for determining the migration of DNA fragments. The resulting pattern should be as accurate as can be estimated for DNA molecules with the exception that there is currently no algorithm that will accurately determine the migration of uncut plasmids through a gel (( Electrophoresis. 2002 Aug;23(16):2710–9. “DNA electrophoresis in agarose gels: effects of field and gel concentration on the exponential dependence of reciprocal mobility on DNA length” Randolph L Rill 1, Afshin Beheshti, David H Van Winkle PMID: 12210176 DOI: 10.1002/1522–2683(200208)23:16<2710::AID-ELPS2710>3.0.CO;2–0 https://pubmed.ncbi.nlm.nih.gov/12210176/)) (( Electrophoresis. 2002 Jan;23(1):15–9. “DNA electrophoresis in agarose gels: a simple relation describing the length dependence of mobility” Winkle David H Van 1, Afshin Beheshti, Randolph L Rill PMID: 11824615 DOI: 10.1002/1522–2683(200201)23:1<15::AID-ELPS15>3.0.CO;2-L https://pubmed.ncbi.nlm.nih.gov/11824615/)) .
The tool makes designing digests for checking constructs very easy and quick. For example, to check the orientation of a cloned gene, drag a restriction site in that gene to the gel window to view the correct band pattern. Then for comparison, repeat the ligation in the incorrect orientation and drag the site again. You can add a lane for an empty vector too. You can even print out the gel to take into the darkroom as a guide for cutting out a band.
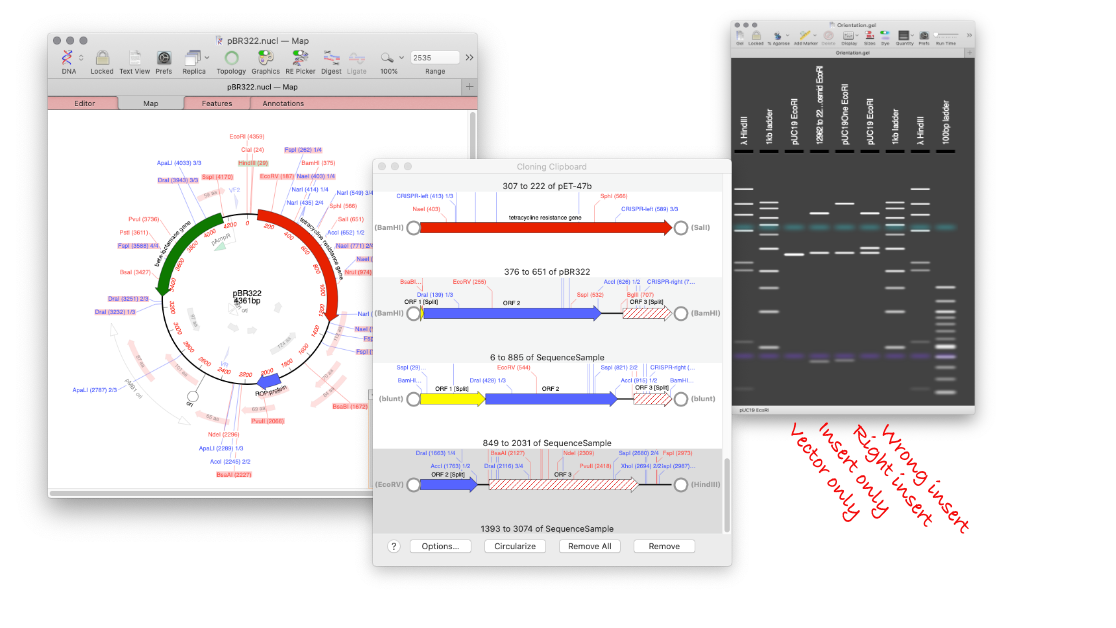
The default settings for the agarose gel display in MacVector generate a reasonable photorealistic simulation of an agarose gel. Although the intensity of small bands has been increased quite significantly, so that they show up a little more obviously and crisply than they might in real life (there’s also no smear from the loading buffer in the wells!). You can select different sets of markers, or define your own. As you mouse-over different bands the status bar updates with details of the fragment(s) under the pointer. The percentage of agarose in the gel and the relative run time can be adjusted as well as the realism of the display. You can optionally view the sizes of the fragments directly on the gel.
The advantages of being able to visualise a gel before you for it are multiple. For example being able to identify which band you need to cut out of a gel before going into the darkroom, or knowing which are the ideal restriction enzymes to cut your mini preps with to produce different banding patterns between a correctly ligated fragment, a reversed one and an empty vector…
How to simulate an Agarose Gel for your DNA sequences.
For a single digest
- FILE > NEW > AGAROSE GEL
- Open your sequence and switch to the MAP tab.
- Drag a restriction site from the Map tab and drop on the Gel window
For a double digest
- FILE > NEW > AGAROSE GEL
- Open your sequence and switch to the MAP tab.
- Select one restriction site, hold down SHIFT and select a second restriction site.
- Drag the sites from the Map tab and drop on the Gel window.
To change the gel marker
- Click the ADD MARKER toolbar button.
- Choose a new DNA marker.
- To remove a lane
- Select the lane.
- Drag and drop the lane outside of the gel window.
