Category: Releases
-
Upgrade to the most feature packed version of MacVector yet and install on multiple Macs with a 50% discount.
Personal licenses are ideal for using on a single Mac, but not if you have multiple Macs or want to install on a shared lab computer as well as your personal Mac. So why not upgrade to a standard license of MacVector Pro 17 that you can share among a group of users in the…
-
What can MacVector do for my lab?
Here’s what MacVector can do for your lab. Comparing sequences Whatever type of alignment your sequence needs, there’s a tool in MacVector. CRISPR Indel Analysis: Identify insertions and deletions following CRISPR editing of a target. Compare Genomes: Compares two related annotated genomes to identify identical, similar and weakly similar features. Sequence assembly of NGS data…
-
MacVector 17: Our most feature packed release yet..
MacVector 17 will be released later this week. Get ready for our biggest release yet. MacVector 17 compares genomes, makes restriction enzyme cloning easier, automates the design of Gibson Assembly and Ligase Independent Cloning strategies. Makes plasmid maps even more beautiful and supports macOS Mojave’s Dark Mode to aid concentration on those late night primer…
-
Scan For… Missing Primers: Automatic Primer Binding Site Display
One new feature in our MacVector 17 release is the ability to automatically display primer binding sites in each DNA sequence that you open. Here’s an example of a couple of primers displayed on the popular pET 47b LIC cloning vector on each side of the LIC cloning site. The image shows how MacVector 17…
-
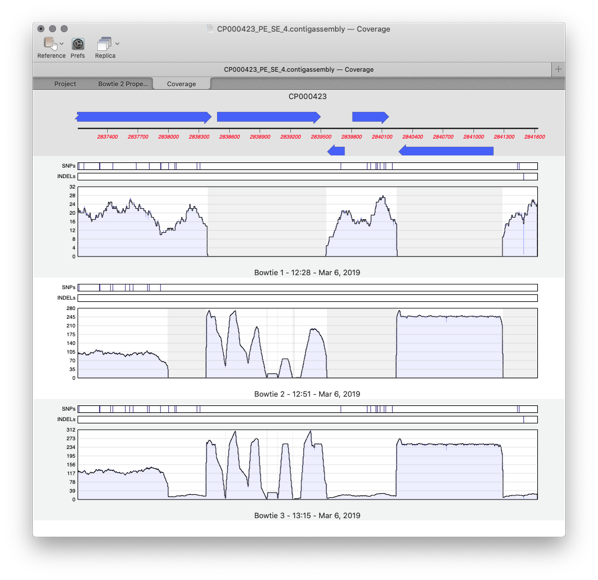
Comparing multiple reference assemblies
MacVector 17 has a greatly improved Assembly Projects manager, for better organization of multiple sequencing datasets, multiple references sequences and repeated jobs. Every time you run a new assembly job (either a reference assembly or de novo). A new job object is created in the Assembly Project window contains resulting contigs and any unaligned reads…
-
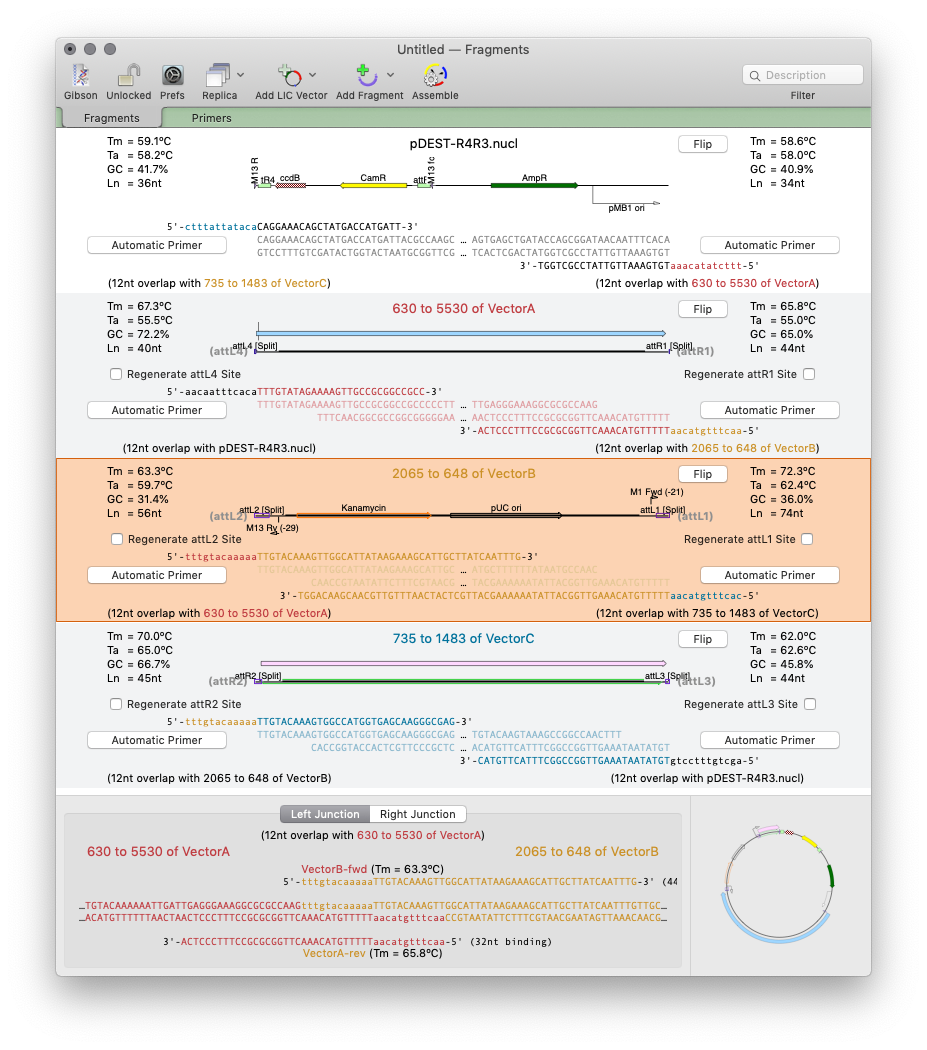
Designing primers for Gibson Assembly with MacVector 17
MacVector 17 has a completely new tool for automated design of ligase-independent cloning strategies. The tool supports 5’ exonuclease driven Gibson assembly as well as the T4 DNA Polymerase 3’ exonuclease “Ligase Independent Cloning” approach. MacVector can automatically design primers when you specify fragments and vectors to use. You can provide custom primers (manually or…
-
MacVector 16.0.10 and BLAST/Entrez issues
We recently became aware of a few issues with Entrez and BLAST. The most common issue is that in Entrez there are only two databases that you can search. The main cause is due to some changes the NCBI made to their service, that went live on the 1st of December, 2018. During the implementation…
-
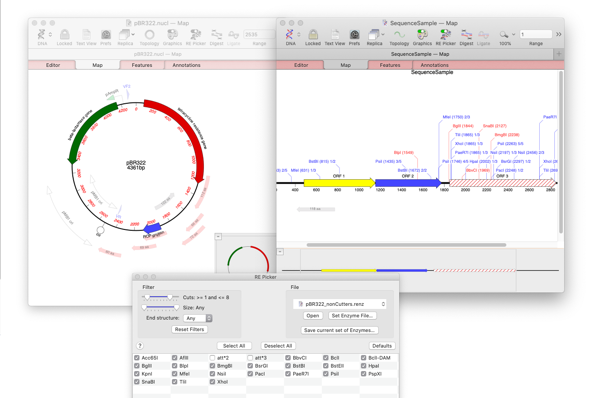
Making restriction enzyme cloning easier with MacVector’s Restriction Enzyme Picker.
MacVector 17’s brand new Restriction Enzyme Picker is a new floating tool for selecting and filtering Restriction Enzymes to simplify the identification of useful enzyme cut sites. It initially presents you with a list of all available sites in a sequence. However, you can filter on many attributes, such as number of cuts, 5’ or…
-
MacVector 16: Our latest release takes automatic sequence annotation to a whole new level
MacVector 16, our latest release, makes beautiful plasmid maps easier, and accurate de novo assembly achievable on your own desktop. Scan for.. Missing Features: Sequences are automatically scanned and missing features displayed. A simple right-click converts them to a permanent feature. Even blank sequences will be displayed fully annotated with common features. You can even…
-
Get ready for the release of macOS High Sierra with a 30% discount on all MacVector upgrades
macOS® High Sierra will be released Monday 25th of September. Whether you intend to upgrade early, or months later, you’ll be pleased to know that MacVector 15.5 (the current version) and MacVector 16 (our upcoming release) are both fully supported and compatible with macOS High Sierra. During October we do have a 30% discount on…
