You can use MacVector to align related DNA or Protein sequences. MacVector uses the ClustalW algorithm to automatically align any number of sequences and provides a sophisticated editor that lets you fine tune the alignments. The alignments can be output in a variety of formats or used to generate phylogenetic reconstructions using a number of built-in algorithms.
Creating Alignments
You can create alignments from sequence already open in MacVector, or by creating an empty alignment and populating it with sequences imported from disk. You can also directly open an existing file containing multiple sequences. MacVector supports the following multiple sequence file formats:
MacVector
PHYLIP
NEXUS
FastA
GCG MSF
GCG RSF
You work with alignments in the multiple sequence alignment editor. Sequences can be viewed using plain monochrome text or colored according to user-defined color groups.
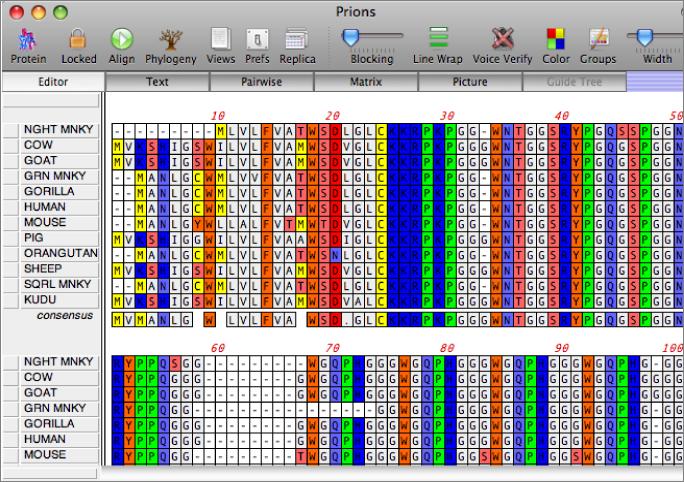
Automatic Alignments
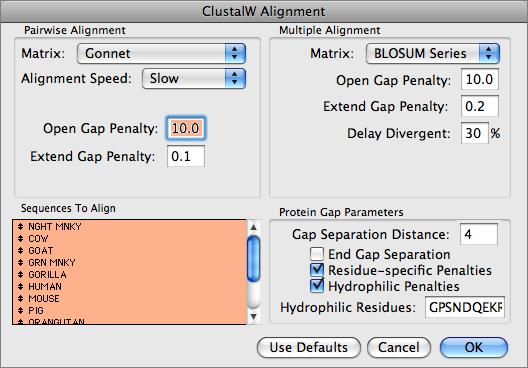
MacVector uses the ClustalW algorithm to align both DNA and Protein sequences. ClustalW 1.83 is used and alignments are submitted into the Job Manager to run in the background, allowing you to continue to work with MacVector.
Editing
The MSA editor provides full editing facilities, including inserting new residues or even adding new sequences from the clipboard. You can change the order of the sequences, copy regions of the alignment to other windows and slide selections around on the screen. The consensus sequence is always updated dynamically and you can control how the consensus is calculated.
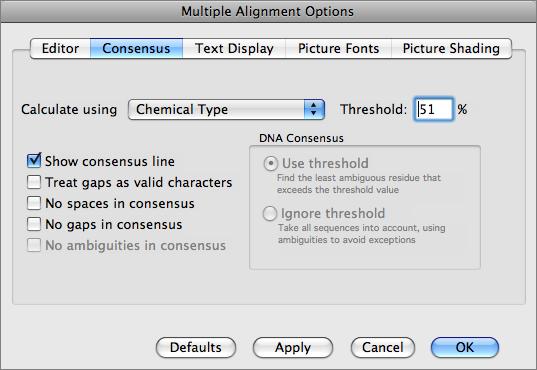
There are a variety of parameters and modes that control how the residues appear on-screen. For example, you can set up the editor so that all matches to the consensus are displayed as dots – this is very useful for identifying variant regions in otherwise closely related sequences.
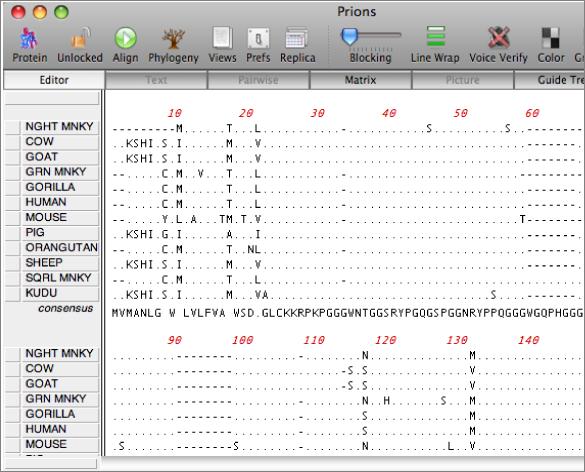
Output Options
The aligned sequences can be displayed in result windows in a variety of different ways. One window provides a high quality customisable graphical text display designed for output on a laser printer, while another duplicates the output from ClustalW. You can also view the sequences aligned in pairwise mode, along with a matrix displaying the identities and similarities of each pair of sequences.
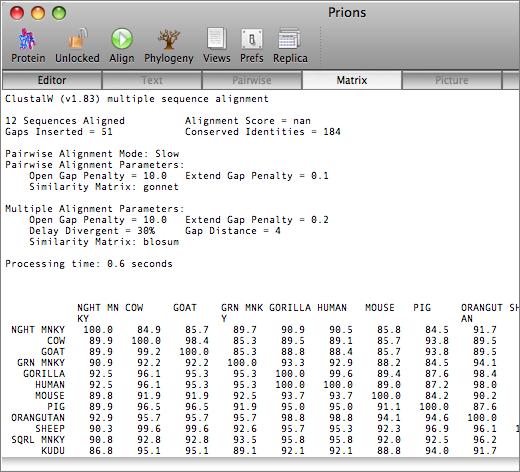
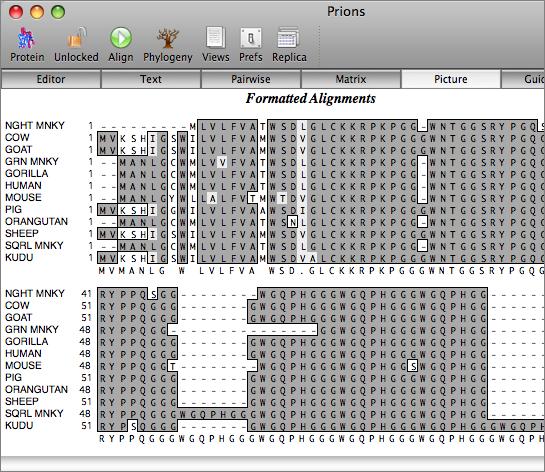
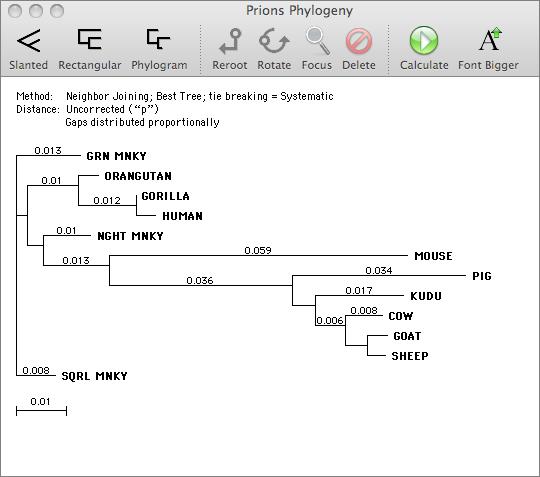
Phylogenetic Reconstruction
You can use MacVector to generate phylogenetic reconstructions from a multiple sequence alignment. Clicking on the ‘create tree’ icon in the multiple sequence alignment editor brings up a phylogenetic reconstruction dialog where you can choose the algorithm and distance correction methods to use. MacVector supports two main reconstruction algorithms – Neighbor Joining and UPGMA, that can be used with Best Tree or Bootstrap calculation modes. A variety of distance correction and gap treatment parameters allow further refinement of the algorithms.
The reconstructed phylogeny is viewed in a graphical window where you can control the type of tree displayed (phylogram or slanted or regular cladogram), the node used for rooting and a variety of other display characteristics.