Assembler
MacVector contains a powerful set of sequence assembly tools which we call Assembler. Assembler was designed from the ground up to be straightforward to use and allows you to manage large amounts of data without yourself without having to send it off to your Bioinformatics department.
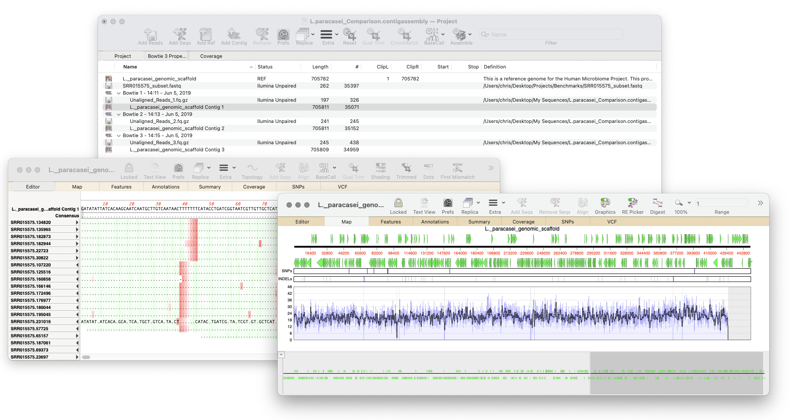
Assembler provides Projects manager and a powerful sequence assembly toolkit. It is easy to use and will manage, assemble and analyze all types of sequencing data – from Sanger reads, all types of NGS sequencing reads, to PacBio and ONT long reads. Assembler includes six different de novo assemblers, as well as two assemblers for mapping reads to reference sequence.
- Align to Reference – cDNA Alignment
- Align to Reference – Sequence Confirmation
- Assembling Sequences Using phrap
- Base Calling Using phred
- Creating a Sequencing Project
- de novo NGS Assembly Using Velvet
- Editing and Analysis of Contigs
- Functional Comparison with Sequencher
- Long-Read Reference Alignments using minimap2
- NGS Reference Assembly Using Bowtie
- Trimming Vectors
