Comparing MacVector Pro with Assembler with Sequencher
Sequencher doesn’t run on macOS 10.15 (Catalina) and later versions of macOS
If you use the popular Sequencher sequence assembly program from Genecodes you cannot run the current version of Sequencher
The root of this problem is that Sequencher 5.4.6 (the latest version for the Mac) is a 32-bit application and is particularly reliant on an ancient “Carbon” OS 9-compatibility framework that Apple has been telling developers for many years will be phased out in the near future. That day finally arrived and until Sequencher is completely rewritten for the latest macOS, you cannot upgrade your Mac to Catalina.
MacVector with Assembler has been fully 64-bit for many years (since MacVector 13) and has no dependency on the “Carbon” framework. It provides almost all of the functionality of Sequencher within an updated modern interface, along with a host of additional functionality and integration with all of the DNA analysis tools within the core MacVector application.
For a limited time, we are offering a 50% discount to all Sequencher users who wish to upgrade to MacVector with Assembler.
Project Interface
Like Sequencher, MacVector with Assembler provides a simple Project-oriented interface where you can import and drag and drop files for Assembly. Unlike Sequencher, MacVector lets you have multiple projects open at one time.
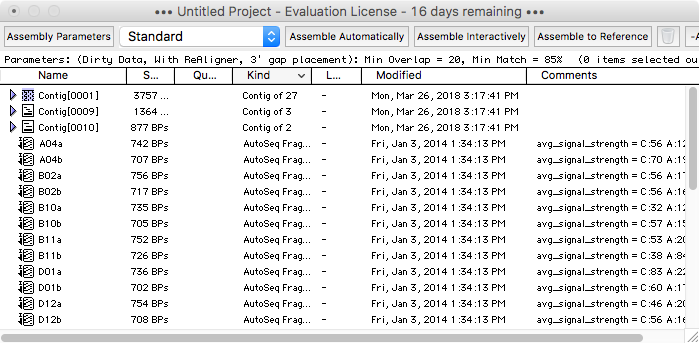
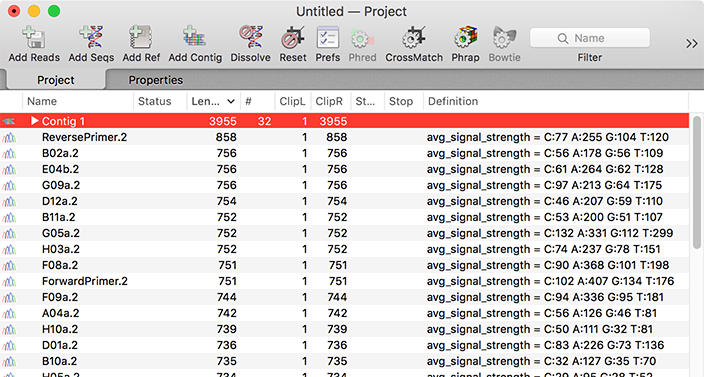
Viewing and Editing Contigs
Sequencher uses two coordinated windows to displaythe contig alignment and any associated chromatogram files;
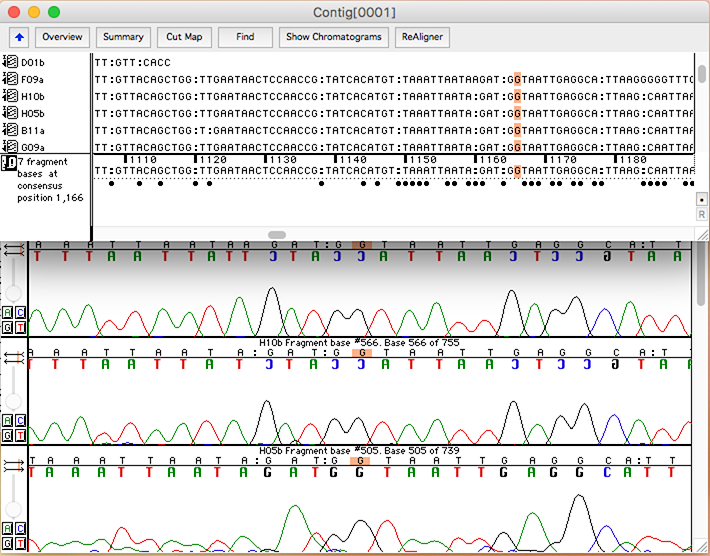
MacVector displays the information in two panes in a single window, along with (optionally) quality information. Unlike Sequencher, trimmed bases are not deleted, but are simply shown greyed out and can be hidden/shown with a simple toolbar button click.. A unique “Dots” mode (not shown) replaces residues that match the consensus with a dot so that your eyes are immediately drawn to SNPs and other mismatches.
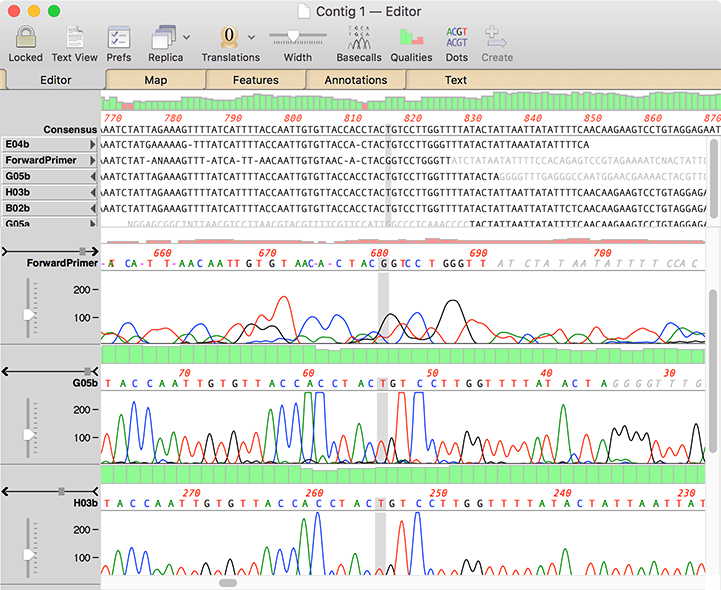
Comparison of MacVector Pro with Assembler and Sequencher Functionality
While not every Sequencher function and feature is present in MacVector with Assembler and MacVector provides many additional sequence analysis functions that are not present in Sequencher, the two applications are broadly similar when it comes to de novo and reference-based assembly capabilities. The table below list the major features present in the two programs;
| MacVector | Sequencher | |
| Assembles ABI Data | Yes | Yes |
| Assembles NGS Data | Yes | Yes |
| Simple Project Interface | Yes | Yes |
| de novo Assembly | Phrap, Velvet, SPAdes, Flye | Sequencher, Velvet |
| Base calling with Quality Values | Phred | Sequencher |
| Reference Assembly | Bowtie, MacVector | Sequencher, BWA, GSNAP, Maq |
| Built-in Reference Assembly Editor/Viewer | Yes | Requires external Tablet viewer |
| Supports Quality Values | Yes (standard phred values) | Yes (Sequencher) |
| SNP Detection | Yes | Yes |
| Multiple Sequence Alignment | ClustalW, MUSCLE, T-Coffee | ClustalW, MUSCLE |
| Restriction Enzyme mapping | Extensive | Minimal |
| Variant calling with SAMtools | Yes | Yes |
| Multiplex ID | No | Yes |
| Modern graphical interface | Yes | No |
| High quality vector-based graphical export | Yes | No |
| 64-bit | Yes | No |
| Compatible with future macOS releases | Yes | No |
| Scan fasta/q files and retrieve matches | Yes | No |
| Built-in NCBI BLAST | Yes | No |
| Extensive DNA analysis and editing tools | Yes | No |
| Save unaligned reads to separate file(s) | Yes | No |
