MacVector 18.8 is the latest release of the best sequence analysis application for the Mac. MacVector 18.8 is a Universal Binary application and will run natively on both Apple Silicon Macs and Intel Macs.
We think there is no better tool for designing primers, preparing beautiful plasmid maps, comparing sequences, CRISPR INDEL analysis, Gibson Assembly, LIC and restriction enzyme cloning workflows, comparing genomes, sequence assembly and much more on the Mac.
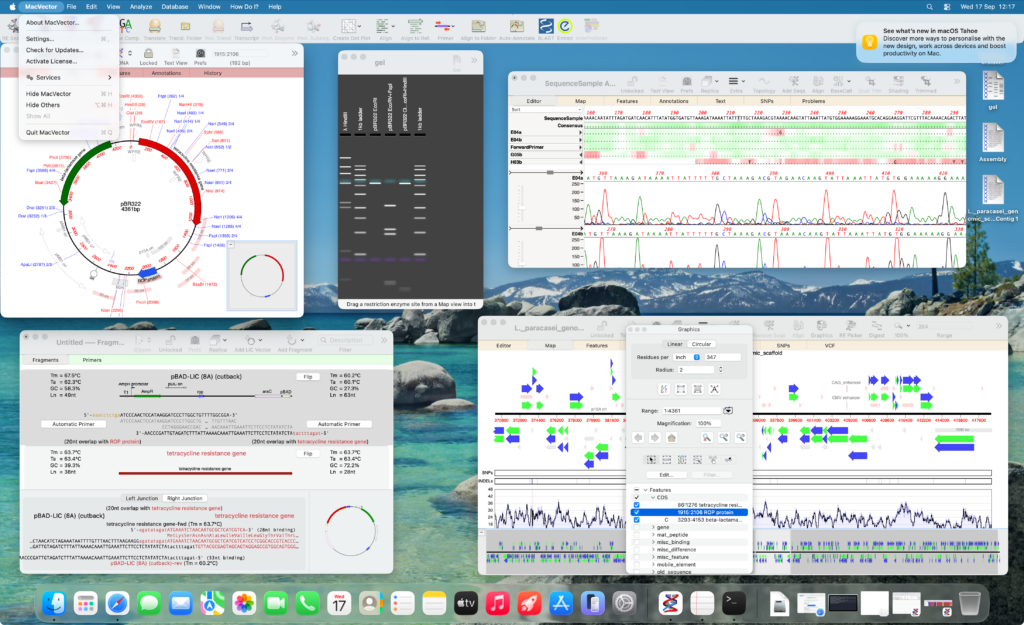
MacVector 18.8 is fully supported on macOS Tahoe to macOS Big Sur.
Design Cloning workflows
As simple as dragging a fragment to a cloning vector.
Cloning history: Track the history of your sequence in the History tab.
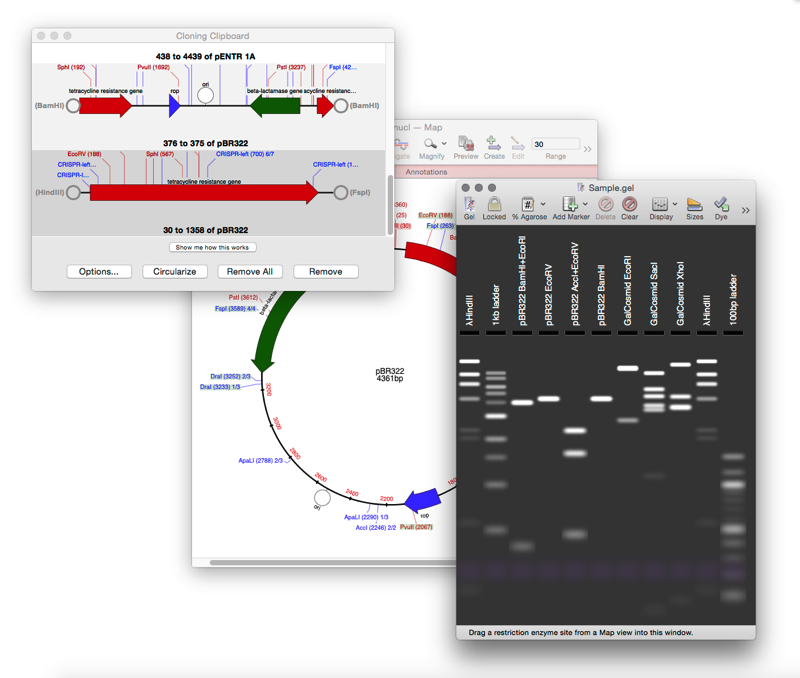
Cloning Clipboard: Store your digested fragments ready for cloning.
One step codon usage optimization: for enhanced expression in different organisms.
Custom Codon Usage Tables: Generate your own codon usage tables or import new ones.
Simulated Agarose Gel: identify restriction sites to screen minipreps.
Restriction Enzyme Picker: easy identification of useful enzyme cut sites.
Gibson/Ligase-Independent Assemblies: project based design of workflows inc. primer design.
QuickTest Primer makes primer design easy! Slide your primer along the template until it aligns perfectly!
Add tails to your primers with silent restriction sites/mismatches and view reading frame changes.
Scan For… Missing Primers: automatically display binding sites from your lab’s primer collection

Easily create beautiful plasmid maps
Automatically annotate missing features, primer binding sites CRISPR PAM sites and putative ORFs.
InterProScan: Scan proteins for functional domains.
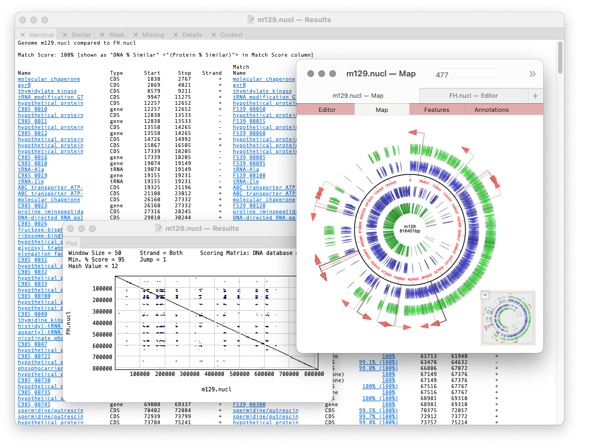
Compare Genomes: identify identical, similar and weakly similar genes.
CRISPR Indel Analysis: following CRISPR editing of a target.
de novo sequence assembly using Phrap, Velvet and SPAdes with Flye for PacBio and Oxford Nanopore.
Reference sequence assembly using Bowtie2 and Minimap2 for PacBio/ONT long reads.
Directly import Sequencher assemblies into MacVector’s Assembly Projects.
Sub-project assemblies: Assemble reads into sub-projects in a single assembly.
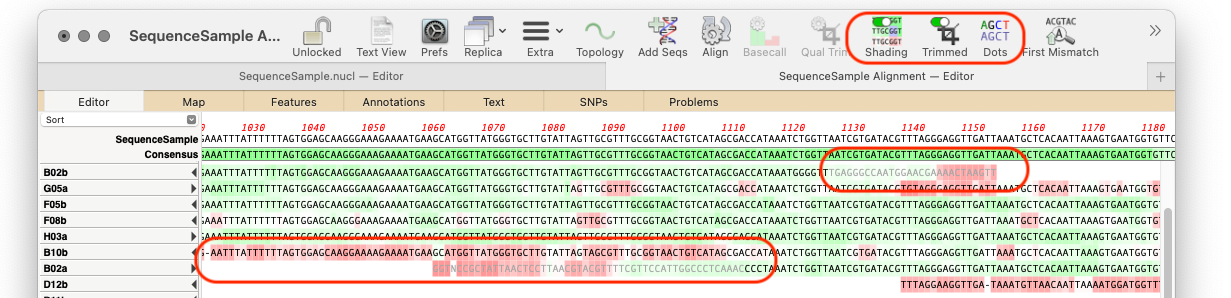
Sequence confirmation: quickly verify sequenced constructs.
Heterozygote analysis: report on possible heterozygotes and represent with IUPAC ambiguity codes.
Coverage Tab: compare expression level across assemblies.
RNA-Seq analysis: Read depth visualization with per gene coverage data (RPKM & TPM).
Batch BLAST automates BLAST searches across datasets. For example identifying contigs from an unknown sequenced samples.
Circularize genomes: toolkit for finishing assemblies and circularizing bacterial ones.
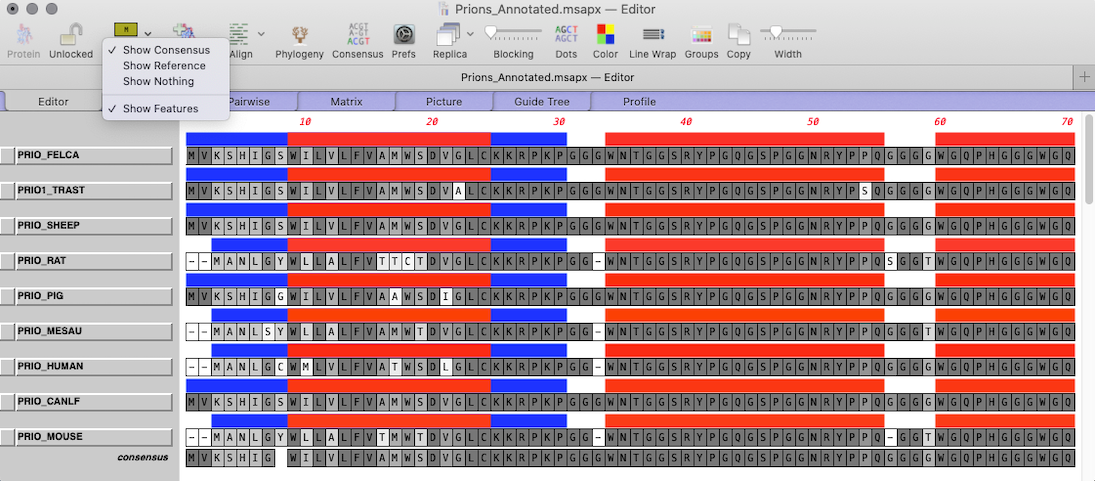
Outline shared aligned domains: Multiple sequence alignments display shared domains.