Tag: phrap
-
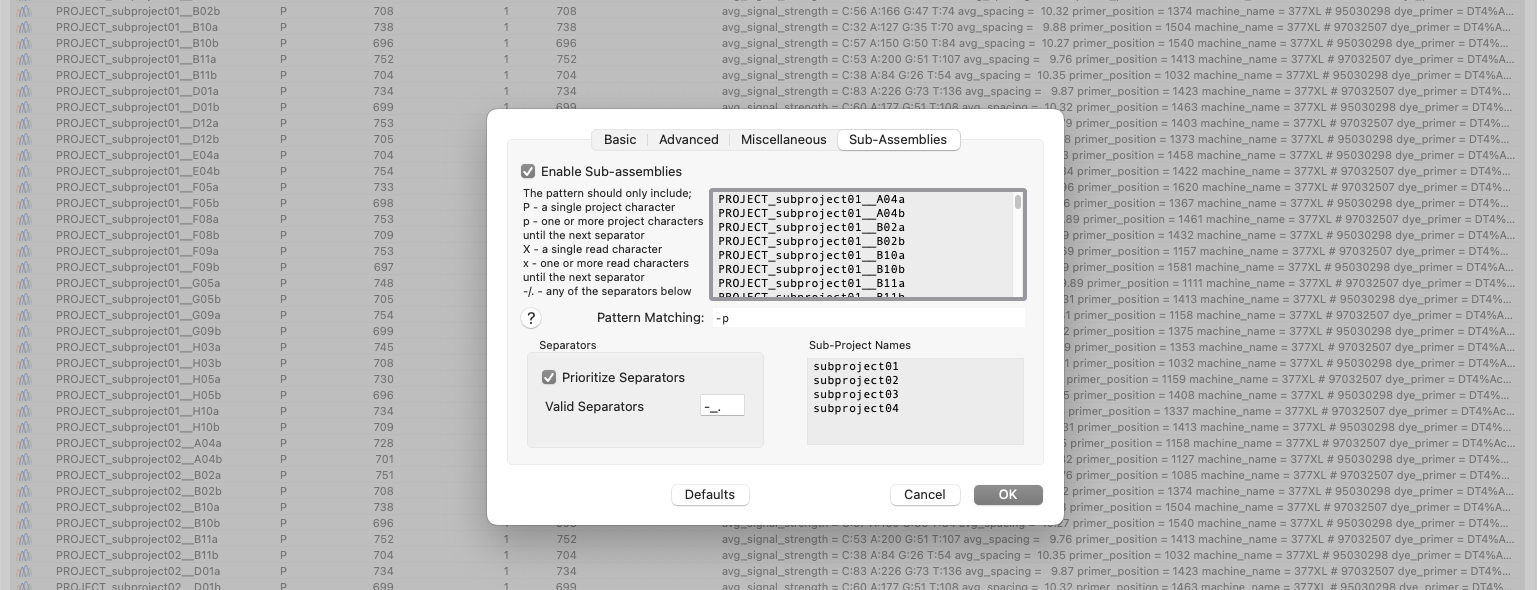
Automatic Assembly of Sub-projects with Phrap (Sub-Assemblies)
New to MacVector 18.6 is the ability to sort and assemble reads from different datasets into individual sub-projects. This functionality is located in the phrap parameters dialog. When enabled and configured appropriately for your dataset it will automatically break out the input reads into sub-projects to be assembled separately. A simple pattern-matching text box lets…
-
Assembling sequencing data with MacVector and Assembler
Update: As of September 2024 Assembler will be added at no cost when you upgrade or renew maintenance. Contact Support if you have not yet been upgraded. To assemble various types of sequencing reads, follow these steps. Then follow one of the following: To create a de novo assembly from Sanger reads To create a…
-
How Assembler uses quality scores to create assemblies
A common problem with all types of sequence assembly is distinguishing between sequencing errors and true genomic variations. Quality scores are one way to help the algorithm identify if a variation is of high quality and therefore likely to be a SNP or a sequencing error. For Assembler trace files can be basecalled with Phred,…
