Tag: sequencing
-
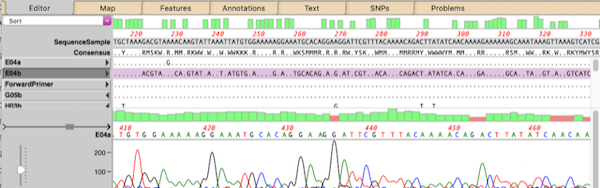
MacVectorTip: Advanced Align to Reference Editing
You can use the Analyze | Align to Reference function to align other sequences (Sanger chromatograms, plain sequences or even NGS data collections) against a reference. Once aligned, the Editor lets you perform all the usual editing functions using an “overwrite” mode – select the residue you want and type the new residue to replace…
-
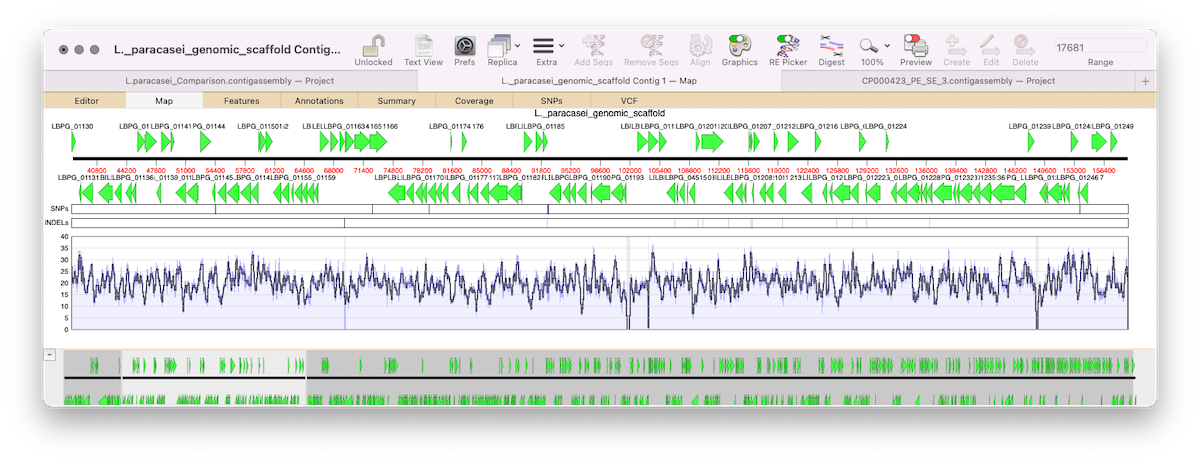
Importing Sequencher project files into MacVector
Assembler is a plugin for MacVector that provides comprehensive sequence assembly functionality. Assembler is fully integrated into MacVector and allows you to manage sequencing data with the familiar MacVector style. You can design primers directly on a contig or BLAST that contig to identify it. MacVector’s Assembly Project manager Like Sequencher, MacVector with Assembler has…
-
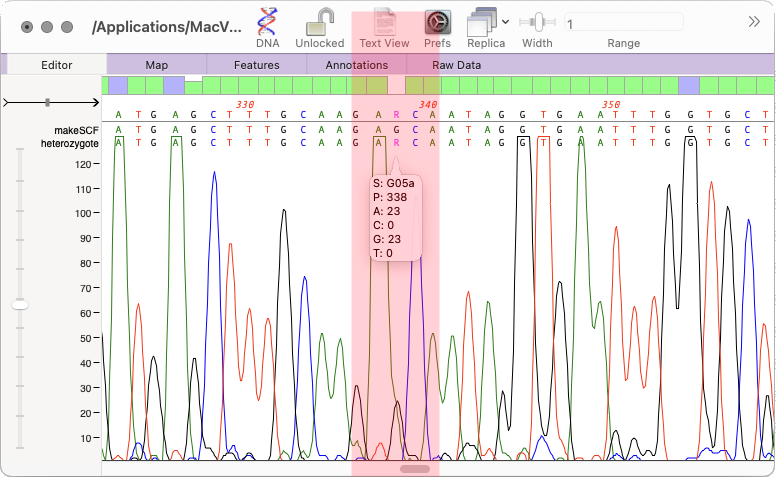
How to call heterozygotes in trace files or Assembly Projects
In MacVector 18.5 we added a new tool to call heterozygotes in sequencing reads. The heterozygote analysis tool allows you to either view heterozygotes in Sanger trace files or to permanently change the basecalled sequence with an ambiguity representing the called heterozygote. The tool works on multiple trace files in the Assembly project manager or…
-
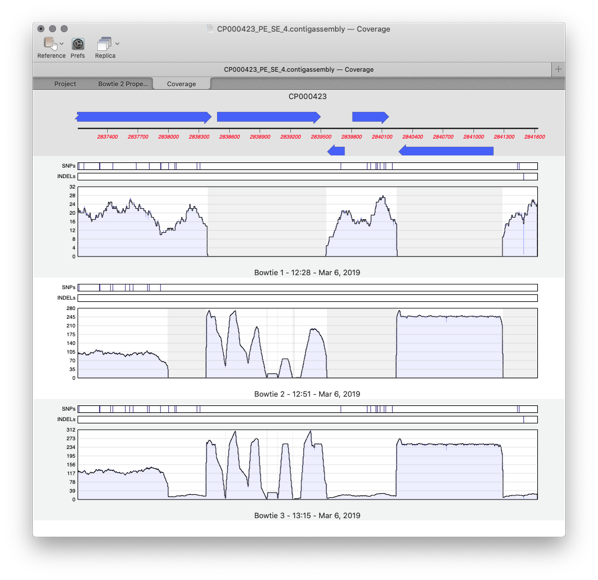
MacVectorTip: correctly flagging PacBio and Oxford Nanopore datasets for assembly by Flye
MacVector 17.5 introduced Flye for assembly of PacBio and Oxford Nanopore reads. Flye joins Phrap, Velvet and SPAdes for de novo sequence assembly using along with Bowtie2 and Align To Reference for reference assembly. Flye is an assembler algorithm tuned to assemble low quality long reads such as those produced by the new generation of…
-
MacVectorTip: quality score visualization in sequence assemblies.
Quality scoring of Assemblies and Align to Reference alignments can be visualized directly on the sequence. Residues can be shaded according to their quality scores. These can be displayed anywhere quality values are available, including de novo and reference assemblies in Assembler and Align to Reference alignments. A Shading toolbar button lets you turn on…
-
Generating a primer report to send to your Oligo Synthesis service
QuickTest Primer is a great tool for primer design. Paired with Primer Design/Test (Pairs) it gives you great control and flexibility for designing primers with tails, mismatches, silent mutations, one out sites and more. Once you’ve designed your primer the next step is to get it synthesized. QuickTest Primer will produce a PDF report of…
-
Confirming a small sequencing project against a reference sequence
Align to reference is a perfect tool for mapping small sequencing projects against a reference sequence. It’s perfect for accurately and quickly:– Confirming the sequence of a cloned fragment– Sequencing across the ends of a cloned fragment to confirm the junction sequence– Screening clones from a site-specific mutagenesis experiment to identify successful mutations– Screening related…
-
Search fastq files and retrieve matching reads into paired fastq files
The Database | Align To Folder… function is essentially your own personal BLAST search of sequences on your computer, but with the advantage that you can scan fasta/fastq containing millions of entries and retrieve matching Reads into a new file. MacVector 14.5 added an enhancement where you can search paired-end read files and retrieve both…
-
Save trace files as images
You can open ABI chromatogram (“Trace”) files directly in MacVector and analyze them like any other sequence. You can also use them in the Align To Reference and Assembler interfaces, where the actual trace data will be displayed, or in any other function that accepts external sequence files. Sometimes you just want to print out,…
-
Quickly checking a small sequencing project
For analyzing large sequencing datasets you need Assembler. However, many times you do not need a powerful tool but just a quick way to check some sequencing data. For example for checking small sequencing projects, such as a site directed mutagenesis, looking for SNPs in a PCR product, cloning a gene or checking your latest…
