Tag: weeklytip
-
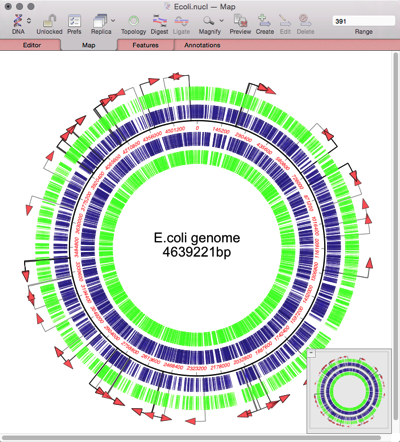
MacVectorTip: Assembling Fungal Genomes using SPAdes
MacVector with Assembler can assemble bacterial genomes in just minutes on quite modest hardware. Currently MacVector has four de novo assembly tools (SPAdes, Velvet, Flye and Phrap). But what of larger genomes? It is currently impractical to run de novo assemblies of Human genomes on a low cost Mac, though RNA-Seq analyses against the human…
-
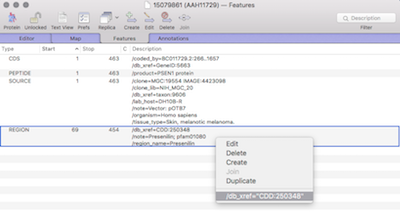
MacVectorTip: Viewing external database entries for features in a sequence.
Sequences, or regions of sequences, can be linked to external databases. For example an entire sequence entry or for when annotation tools are used to annotate proteins with domain or motif information (for example InterProScan). Very useful for when you want to view more detailed or updated information. Within the Genbank specification, which MacVector extensively…
-

MacVectorTip: How to Customize Window Button Toolbars
Like many Mac applications, MacVector takes full advantage of the built-in ability to add, delete and rearrange the action buttons on window toolbars. To make these changes, right-click (or [ctrl]-click) in the gray space on any toolbar and a context-sensitive menu will appear. Choose Customize Toolbar and a dialog will be displayed with all of…
-

MacVectorTip: Understanding Color Groups
You can align hundreds, or even thousands of protein sequences within MacVector using three different alignment algorithms – ClustalW, MUSCLE or T-Coffee. Once aligned, you may be familiar with the colorful display in the Editor tab. But there’s more to this than pretty colors. The default Color Group in MacVector is one called “Chemical Type”.…
-
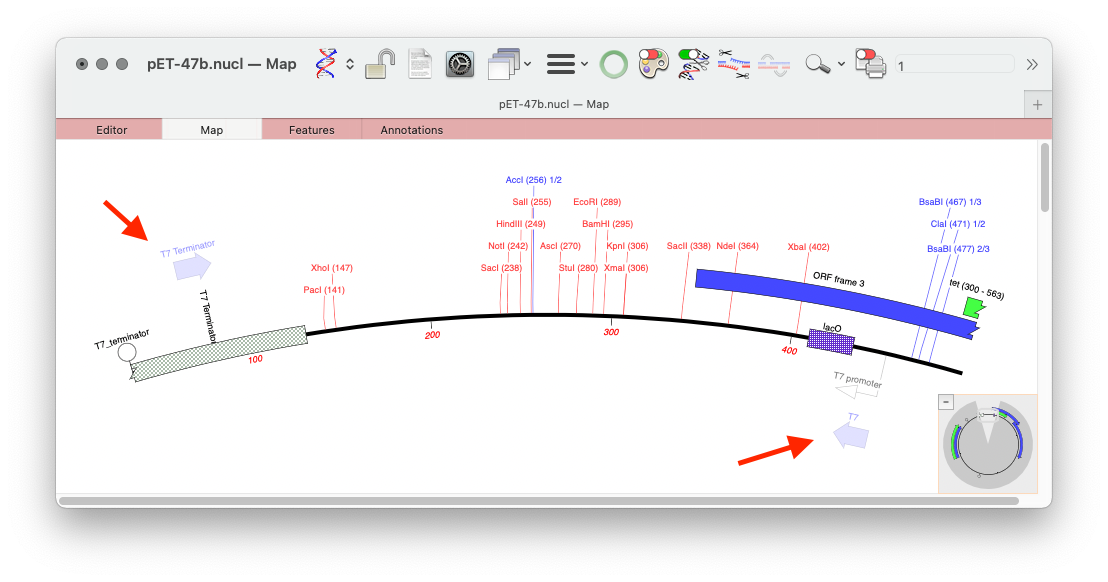
MacVectorTip: Scan For… Missing Primers: Automatically display Primer Binding Site on your sequences
MacVector’s Scan DNA For.. tool allows you to automatically display restriction enzyme recognition sites, putative ORFs, CRISPR PAM sites, missing annotation and also it will display primer binding sites from your own Primer Database in each DNA sequence that you open. Here’s an example of a couple of primers displayed on the pET 47b LIC…
-
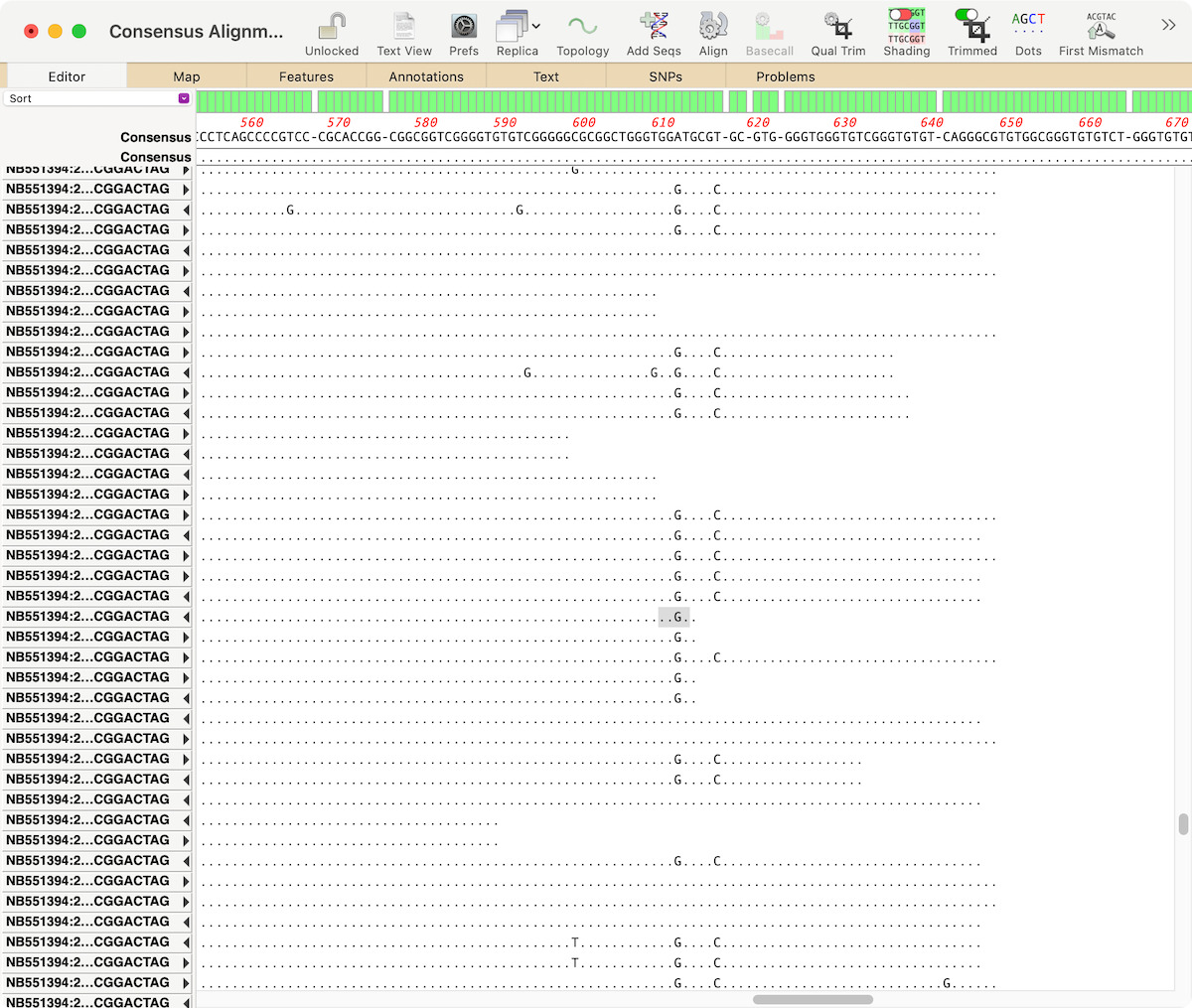
MacVectorTip: Viewing genotype changes in Align to Reference assemblies
The latest releases of MacVector, MacVector 18.0.1 (Intel) and MacVector 18.1.1 (Intel and Apple Silicon) have some tweaks to the output of the SNPs tab in the Align to Reference assembly window. The genotypes of any SNP changes now follow a consistent standard, and short deletions are also reported. If the region containing the nucleotide…
-
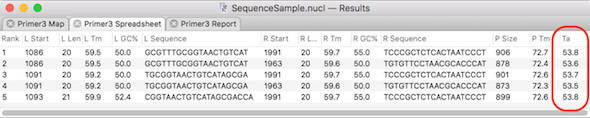
Calculating the optimal PCR annealing temperature
MacVector has several tools to help with primer design and testing. The Analyze | Primer Design/Test (Pairs) function uses the popular Primer3 algorithm to find suitable pairs of primers to amplify specified segments of DNA. You can also enter pairs of pre-designed primers and test their suitability for use in PCR. In both cases, the…
-
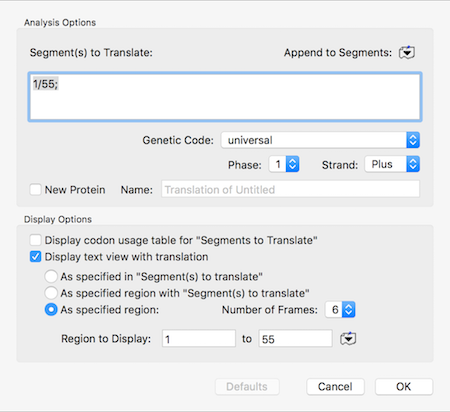
How to copy a specific short amino acid translation of a sequence
There can be times when you are messing about with open reading frames, inserting residues to change frames to try to get the perfect CDS fusion. The MacVector single sequence Editor will show those (click and hold on the “Display” toolbar button) but if you select and copy, only the DNA sequence (with any overlapping…
-
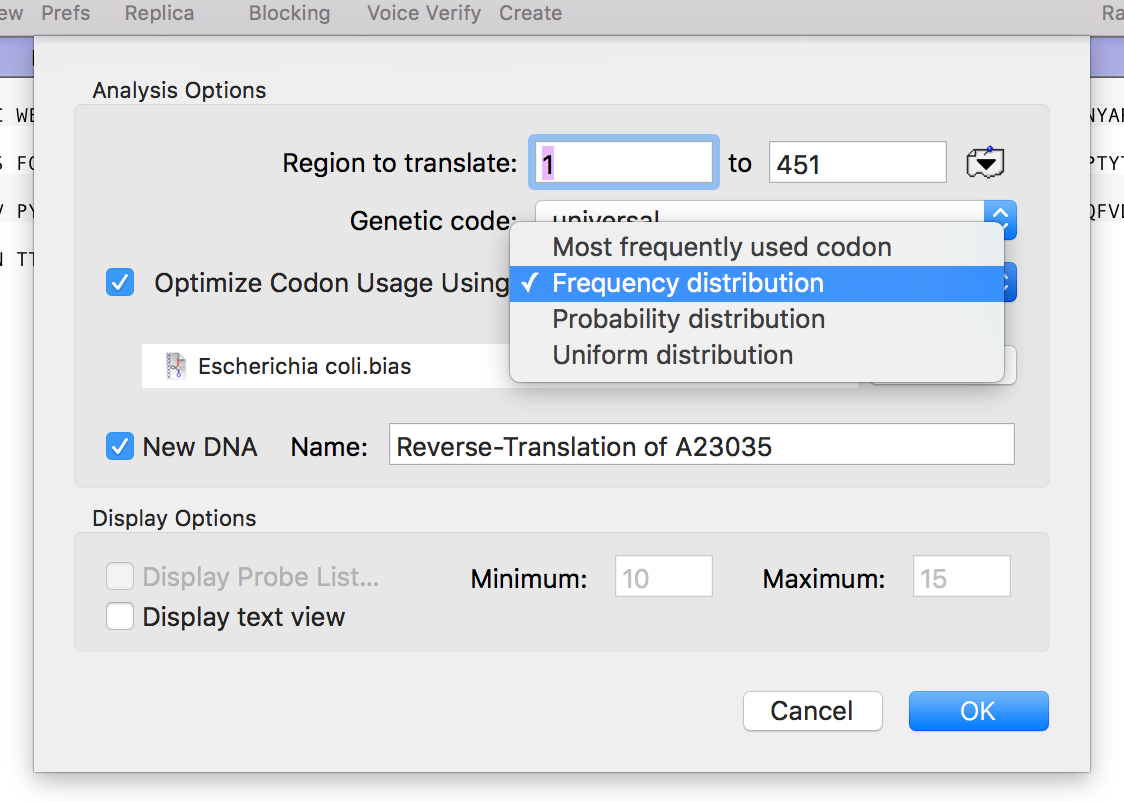
Optimizing Reverse Translations
The Analyze | Reverse Translation menu option lets you create a DNA sequence from a Protein sequence, reverse translated using a specific Genetic Code (by default, the Universal Genetic Code). The default option creates a DNA sequence with N’s and other ambiguities reflecting the degeneracy of the genetic code. This is great if you want…
-
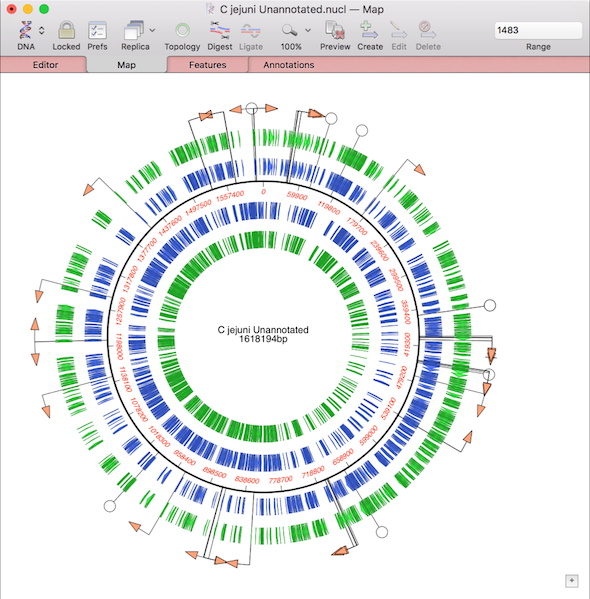
Use Database | Auto-Annotate Sequence to annotate prokaryotic genomes
The continuing advances in Next Generation Sequencing have made it relatively low cost to sequence prokaryotic genomes. Many scientists are embarking on large projects to sequence multiple related genomes. These might be clinical isolates of the same species exhibiting different pathogenetic properties, environmental isolates from different sites, or a study over time of the changes…
