MacVector 18.8
(released July 2025)
Overview
MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Batch BLAST allows you to automate BLAST searches for multiple sequences and Auto-Annotate (via BLAST) allows you to automatically annotate multiple sequences. Both of these tools will run directly from the Assembly Project manager and so are ideal to identify contigs from a de novo assembly job.
MacVector 18.8 was developed on macOS Sequoia and is supported on macOS Big Sur to macOS Sequoia. It will be fully supported on macOS Tahoe when Apple release it (probably in September 2025). MacVector 18.8 is a Universal Binary that will run on Apple Silicon Macs and Intel Macs.
Auto-Annotate (via BLAST)
Auto-Annotate (via BLAST) is similar to Auto-Annotate (local), except instead of using curated sequences on your own Mac (for example the Annotated fragments folder) it uses BLAST to find similar sequences in the NCBI’s BLAST database then annotates your sequence with matching features found in those hits.
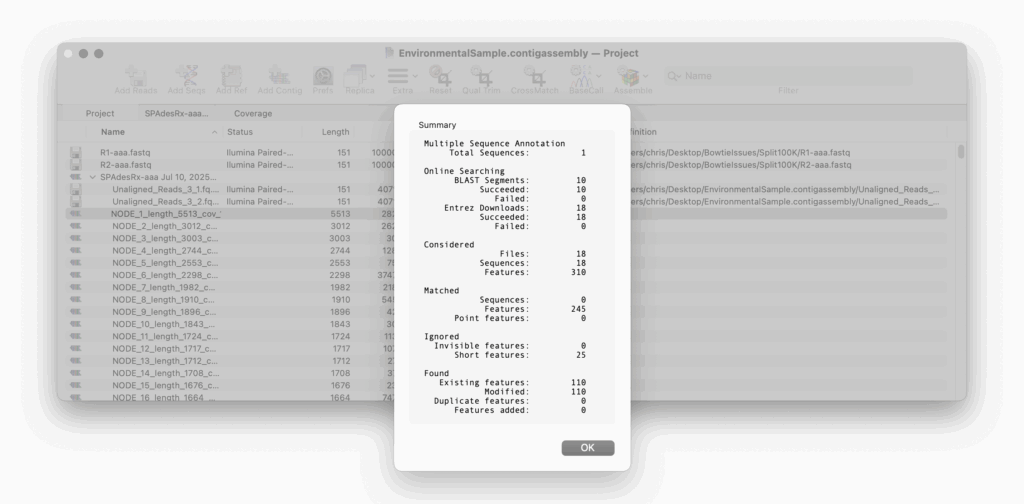
Auto-Annotate (via BLAST) is perfect for annotating an unknown, unannotated or partially annotated sequence. For example a set of contigs from a de novo assembly job.
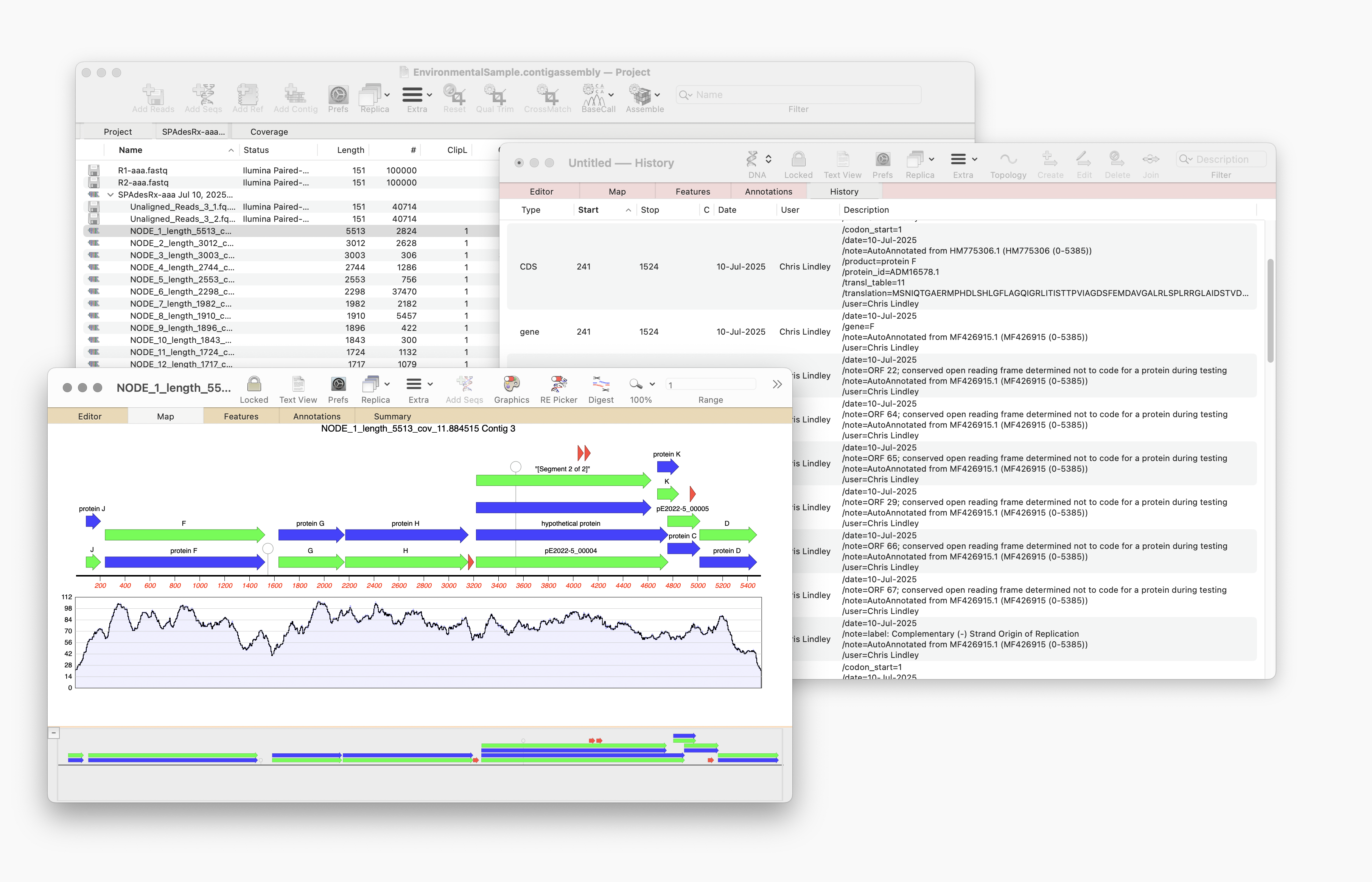
Auto-Annotate (via BLAST) will run on any nucleic acid sequence. However, where it is particularly useful is running directly from the Assembly Project manager to identify contigs that you have assembled. Just select one or hundreds of contigs and run. Every contig will be annotated with features and will aid in identification.
Batch BLAST
Batch BLAST is a game-changing feature that revolutionises the way you identify unknown sequences. With just one simple step, you can identify a whole group of sequence. Ideal for identifying contigs from a de novo assembly.
All you need to do is select your sequence and Batch BLAST will automate the entire procedure of a BLAST search for every one. Batch BLAST will present you with a simple Results spreadsheet with the highest scoring hit for each contig/sequence and will also list any gene (or CDS, mRNA etc.) annotated to the hit sequence.
Batch BLAST is ideal for identifying sequences, for example a bunch of contigs from a de novo assembly job.
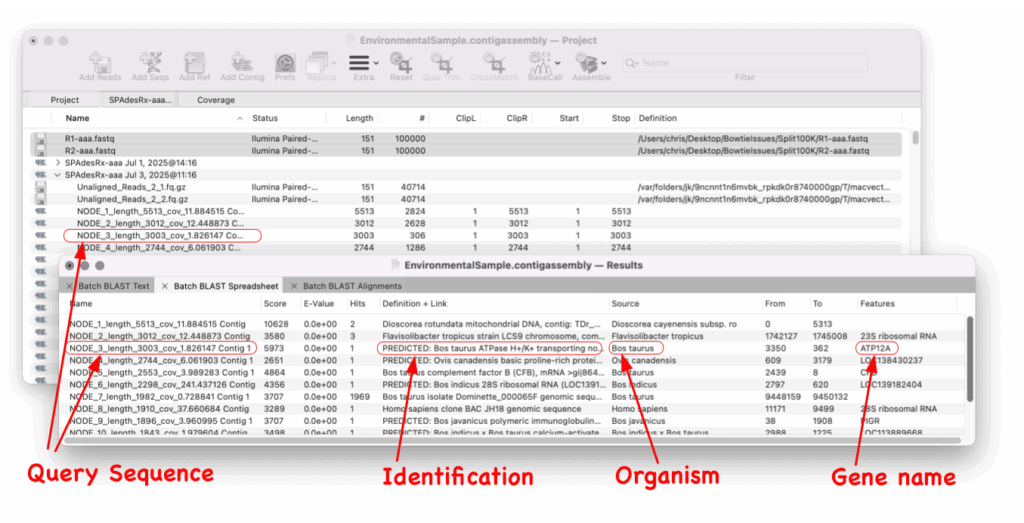
Since Batch BLAST runs directly from the Assembly Project manager you just select the contigs and select Database | Batch BLAST. The results spreadsheet shows the sequence entry and also any gene that is annotated to that hit.
Miscellaneous Enhancements and Bug Fixes
Auto-annotate has been improved to now report and annotate partial feature matches. So, for example, you can analyze a short sequence that matches within a known CDS feature and it will be correctly identified and annotated using the standard GenBank format for “starts before” and “ends after” notation.
There have been a lot of behind-the-scenes changes to the floating Graphics Palette so that it retains its display position and does not scroll when you turn features and Restriction Enzymes on and off.
SPAdes has been updated to a Universal Binary (version 4.2.0).
Bowtie has been updated to version 2.5.3.
A few display bugs in the output of the Compare Genomes tool have been fixed. These particularly affected the alignment of individual matches to the opposite strand or alignments that were not the full length of a feature. These were display glitches only – the underlying data was correct regarding the quality of the match.
You can now assemble pre-designed Gibson Assembly fragments where the end identities were greater than 40 nucleotides.
Align to Reference‘s Map view now displays the clipped section at each end for segmented matches. Previously this was only shown for single segment matches.
The protein pI calculation has been optimized to account for embedded Cys and Tyr residues that rarely practically contribute to the observed pI for a non-denatured protein.
The Multiple Sequence Alignment Editor has been optimized for performance when displaying large alignments.
