In the previous post we discussed the various ways in which you can analyze Oxford Nanopore’s long read data. For de novo assembly we recommend using Flye, which can also be used with PacBio data.
Here are some tips to get the most out of Flye.
IMPORTANT: MacVector simply wrappers around the Flye executable algorithm which depends on Python. Python is included internally within MacVector so you do not need to install it separately. However the current release CANNOT HANDLE SPACES in the filenames of the input reads, or in any of the parental directories where the fasta/q file reside. If Flye exits almost immediately after you dismiss the setup dialog with an error “Flye cannot assemble the reads…” first check that you have no spaces in the “path”. The next release of MacVector will report a more informative error for this problem.
When you invoke Flye, a setup dialog appears;
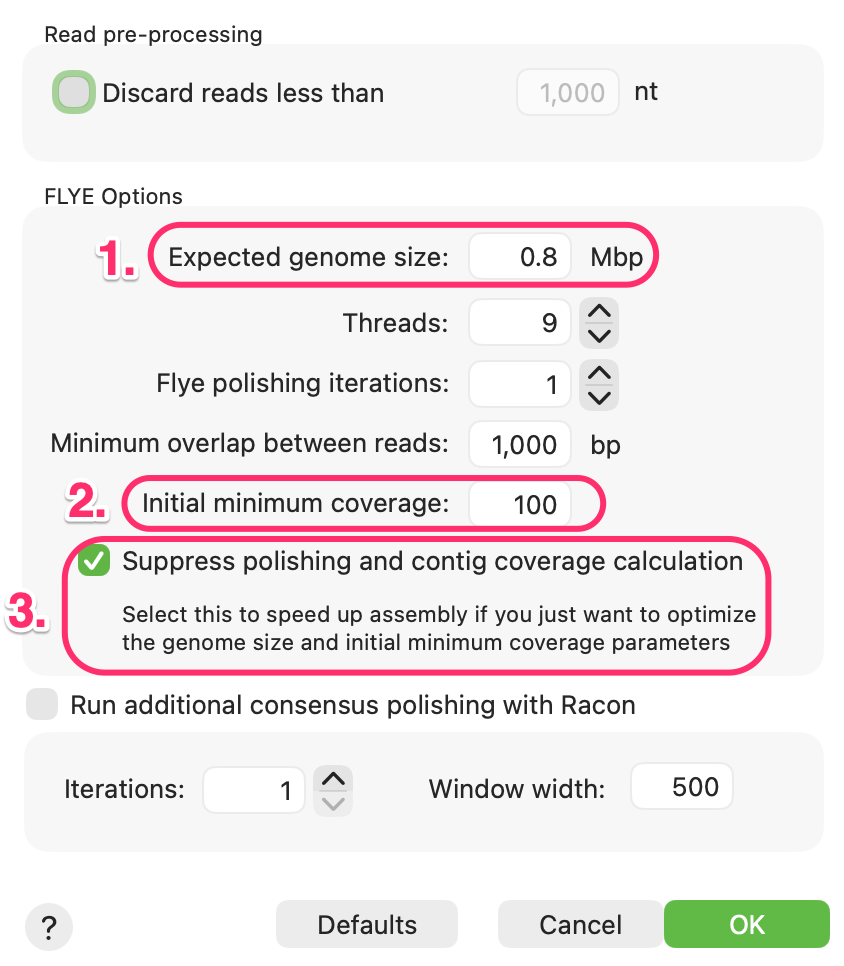
- The most important parameter by far is the Expected genome size: This should be entered in Mbp. It does not have to be perfectly accurate but you should try to get within a factor of 3 or 4 for optimal performance. So, for a 20kb plasmid, you would enter a value of 0.02.
- Flye does not necessarily use all of the data in your input fasta/q files. Instead it will prioritize the longest reads, which also tend to be the most accurate. In this example it will take just enough reads to ensure it will have 100x coverage, so it works in conjunction with the expected genome size to determine exactly how many reads it requires. You can often get faster and more accurate assemblies by reducing this to e.g. 25x as that will prioritize only the longest and (hopefully) best reads in the collection.
- For the most accurate consensus sequence calculations with these noisy long reads, it is recommended to run one or more rounds of “polishing”, either with Flye itself or with the popular Racon tool. However, these can be slow, so it is often a good idea to suppress these calculation while you play with the other parameters to generate the longest and fewest contigs. Once you are happy with your results, you can uncheck that box and select one or more Flye and/or Racon polishing steps and repeat to generate more accurate consensus sequences..
