Author: Chris
-
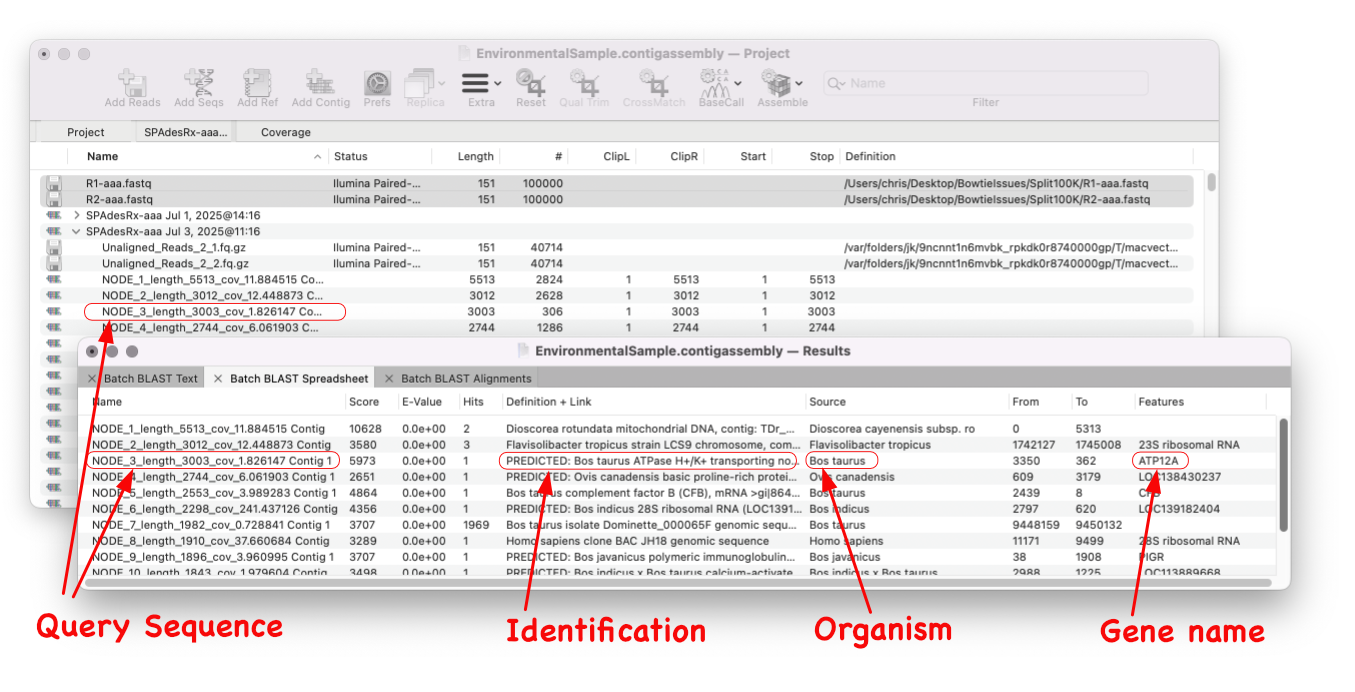
Batch BLAST
MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Ideal for identifying contigs from a de novo assembly. One of these new tools is AutoAnnotate (via BLAST) Batch BLAST is a game-changing feature that revolutionises the way you identify unknown sequences. With…
-
Merry Christmas from the MacVector team
For those who celebrate the MacVector team wish you a very merry Christmas. We hope you are able to turn off that sequencer, put away your pipette and have a relaxing time with your loved ones. Here’s to a better year for science in 2026!
-
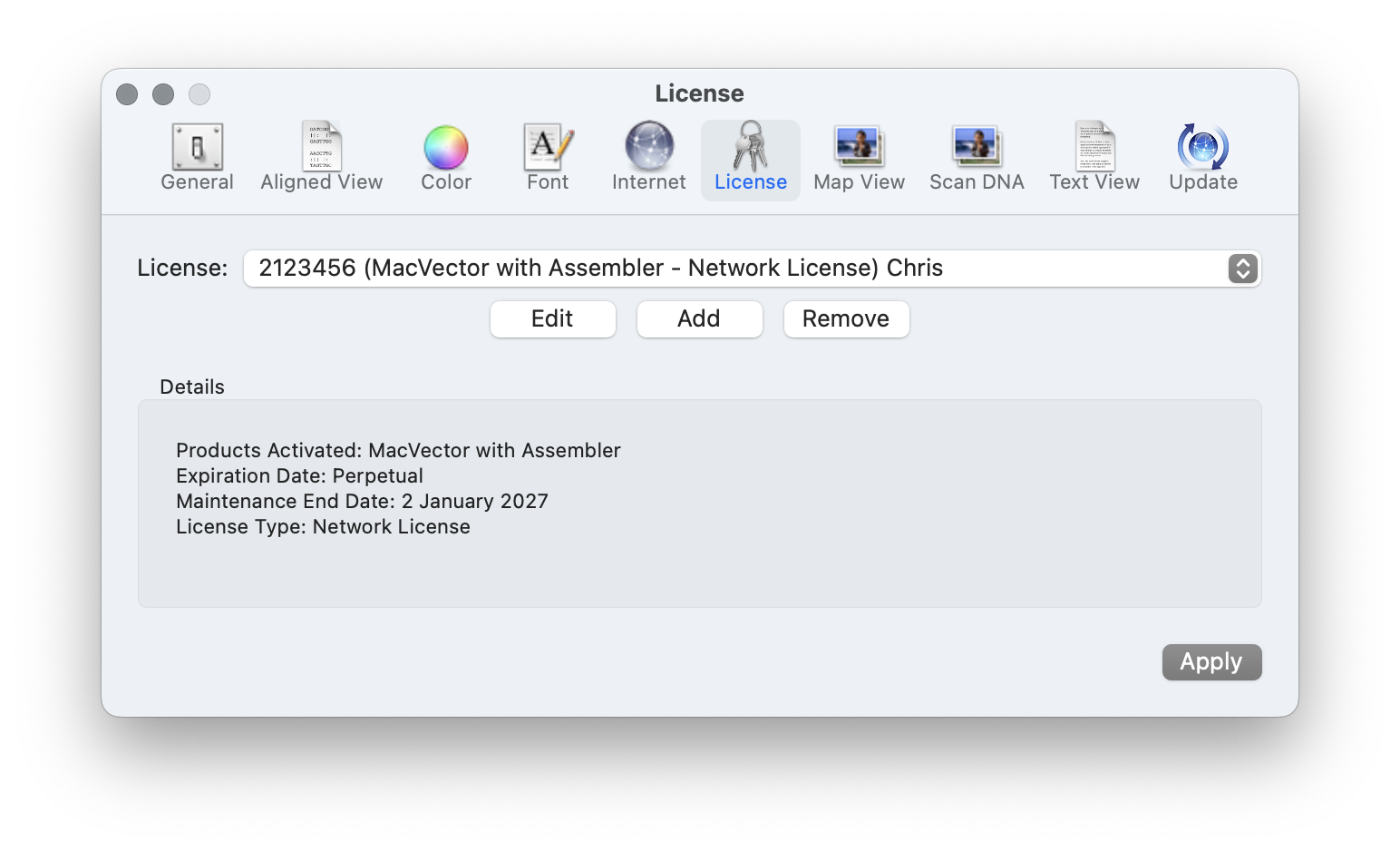
Migrating to a serverless concurrent usage license
As well as having a license per user for larger sites we have concurrent usage or network licenses that allow an entire institute, or a large research group to share MacVector. Concurrent usage licenses give you a pool of licenses that can be shared across an entire network. The number of seats defines how many…
-

MacVector 18.8.2 is out
We’ve just released a minor update to MacVector 18.8. The main change is improvement to Codon Bias Table (CUT) import. But there are multiple bug fixes too. The release notes contain full details. You should be prompted to upgrade in MacVector, but you can also upgrade immediately: MACVECTOR | CHECK FOR UPDATES… You can also…
-
macOS Tahoe and MacVector 18.8
Apple released macOS Tahoe on Monday (15th Sept 2025). As usual in the run up to a new macOS release, we have been testing MacVector 18.8 on development and public betas of macOS Tahoe. With macOS Tahoe’s Liquid Glass appearance we have come across a very minor display glitch. But otherwise we are happy to…
-
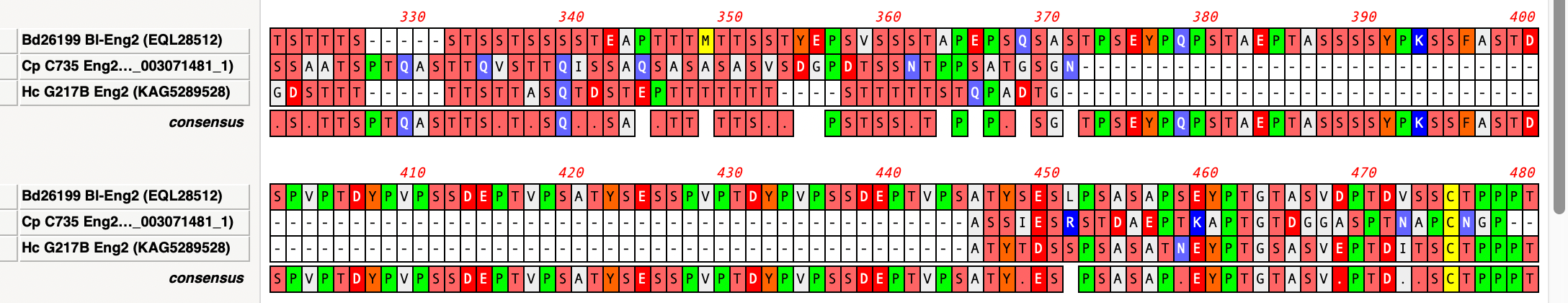
Handling Gaps in Multiple Sequence Alignment Consensus Calculations
By default, MacVector ignores gaps when calculating the consensus of a multiple sequence alignment. However, this can lead to some unexpected results. For example, consider this three sequence alignment where one sequence has a long insertion compared to the other two. In this case it does not seem reasonable to believe that the “consensus” should…
-
Automating annotation of sequences via BLAST
MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Ideal for identifying contigs from a de novo assembly. One of these new tools is AutoAnnotate (via BLAST) Auto-Annotate (via BLAST) is similar to Auto-Annotate (local), except instead of using curated sequences on your own…
-

MacVector 18.8 Is Here – Packed with Powerful New Features
A game changer for identifying unknown sequences MacVector 18.8 is out and it’s packed with new tools! MacVector 18.8 has tools to help you identify and annotate unknown, unannotated or partially annotated sequences. Batch BLAST allows you to automate BLAST searches for multiple sequences and Auto-Annotate (via BLAST) allows you to automatically annotate multiple sequences.…
-
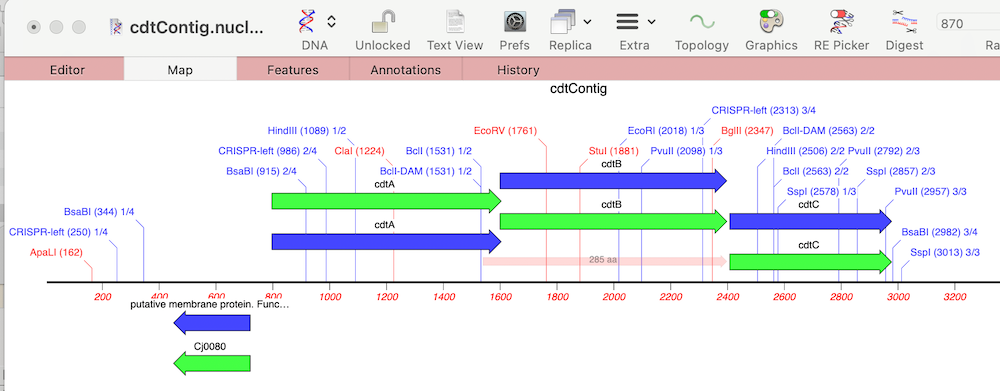
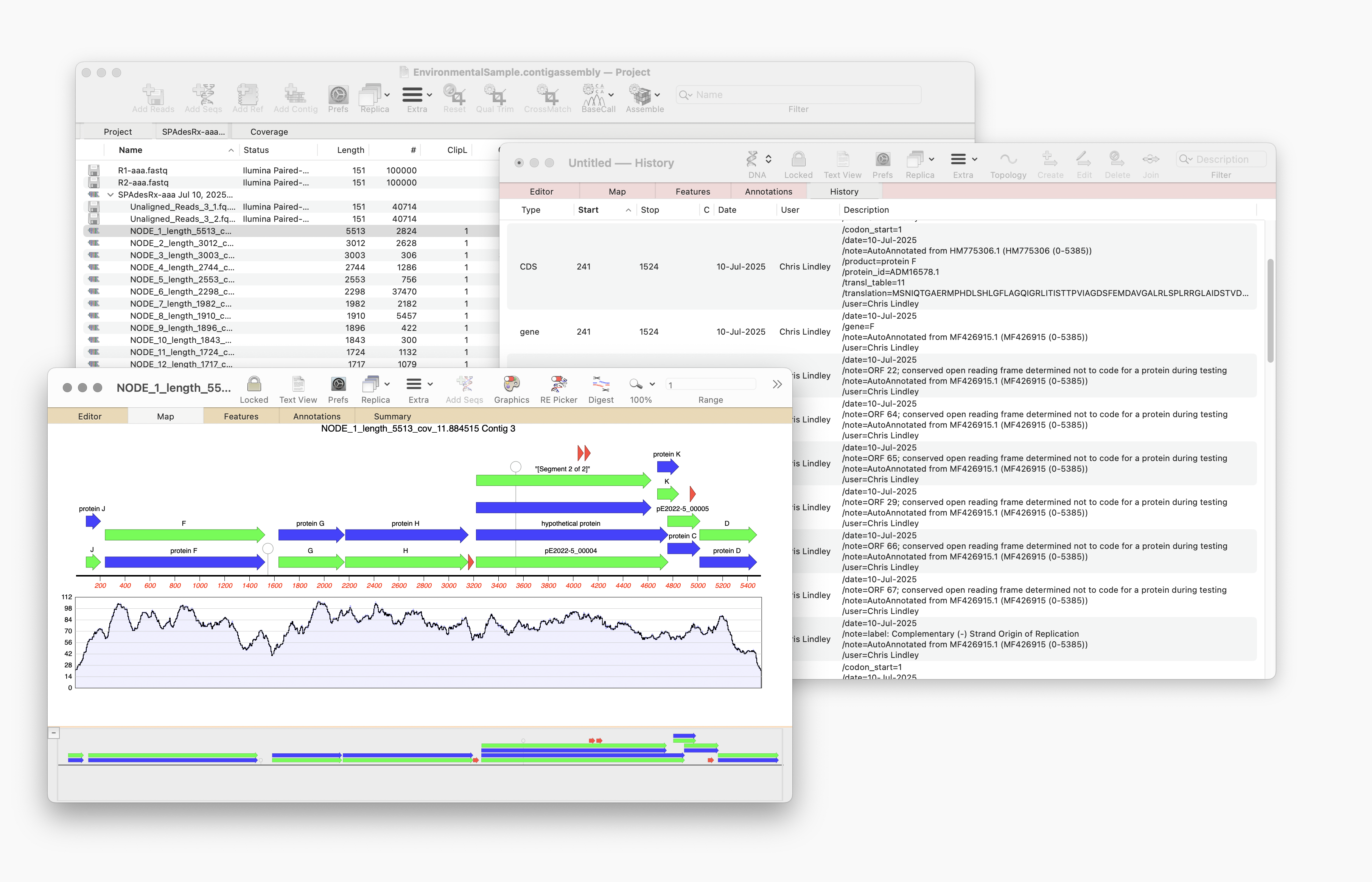
MacVectorTip: Annotating and Comparing Genome Segments
In last week’s tip we showed you how to filter NGS read data to pull out and assemble just those reads that represent a specific gene of interest. Now let’s see how to annotate the single contig we generated and compare that to a reference genome. First, from the Contig Editor, you can save the consensus in MacVector…
-
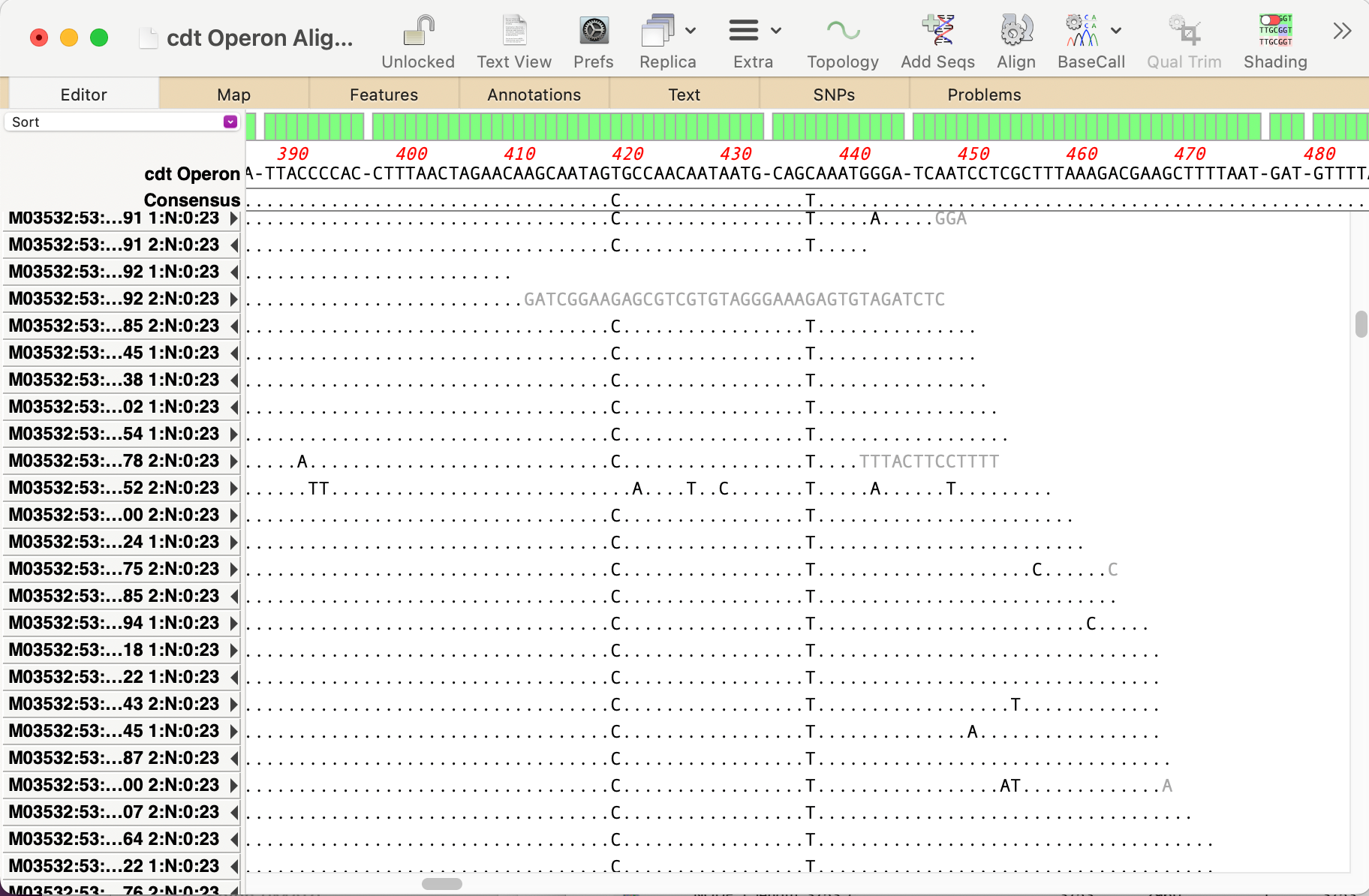
MacVectorTip: Use Align to Folder to filter NGS data for specific genes
Even the latest Macintosh computers loaded with as much RAM as you can afford will still struggle to de novo assemble genomes much over 50 Mbp. But, often that is not required. If you are just interested in a few genes, or a specific region of a chromosome, you can use Align to Folder to filter the…
