MacVector has a brand new tool for designing primers. QuickTest Primer completely changes the way primers can be designed on a computer. It simplifies primer design by showing your primer and its statistics in realtime. Does your primer have a hairpin? Nudge it along your template until the hairpin goes? Want to add a restriction site? Then add one and again nudge your primer to optimise the oligo. Want to see how a CDS translation is affected by your modified primer? View both the original and modified amino acid sequence.
Quicktest Primer can also be used in conjunction with Primer3 to rapidly design pairs of primers with mismatches and/or tails and to generate the predicted product of the reaction, complete with tails and/or mismatches.
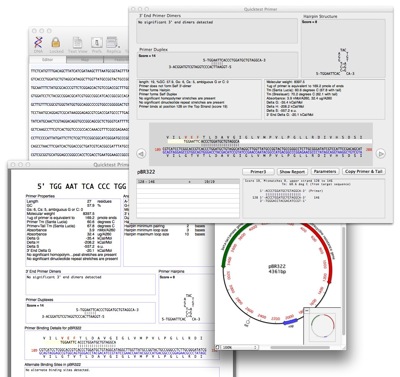
A typical workflow with the new tool would be:
- Design a primer with a mismatch that changes a protein coding region or has a tail containing a restriction enzyme site
- Find a matching primer to amplify a specific segment from a gene and add a restriction enzyme site to that second primer
- Generate a detailed report of your primers
- Generate a new DNA sequence representing the predicted product of the amplification
- Annotate the predicted product back to your sequence.
Quicktest Primer allows you to quickly evaluate the suitability of any short DNA sequence for use in PCR or sequencing experiments. Simply open Quicktest Primer using Analyze | Primers | Quicktest Primer… and type or paste in a primer sequence to display the primer properties instantly. Alternatively, you can evaluate any short portion of an existing sequence quickly by selecting that portion in the Editor, Map or Features view before starting Quicktest Primer.
Quicktest Primer displays a detailed set of properties for a primer sequence and these properties will change in realtime as you modify the primer. For example if you have a hairpin you can nudge your primer along the template until this disappears or if you have added a restriction site you can slide the primer along until it does not change the translation of any CDS feature that your PCR product would contain. QuickTest Primer is linked to the active nucleic acid sequence window. If you select a different nucleic acid sequence window, then MacVector will recalculate the primer properties. Since no binding information is displayed if the selected primer does not bind to the current sequence, you can use this feature to establish quickly if (and where) the primer binds to any open nucleic acid sequences.
Quicktest Primer displays the following information:
- Melting temperature
- Presence of “bad” residues like non-G/C 3’ end and runs of >3 homopolymers.
- Graphical display of hairpin loops and primer dimers.
- Secondary structure scores.
- Location of any other matches on the target sequence.
- Translations of any CDS features that overlap the binding region are also shown.
In summary, Quicktest Primer allows you to:
- Add a user defined 5’ tail.
- Nudge the primer along the target sequence to find the optimum sequence.
- Find a balanced second primer using Primer3
- Create a mismatch at any location (especially corresponding to a “one out” site).
- View secondary structure in real time as you enter a primer sequence.
- Generate a detailed report of your primer.
- Slide your oligo along your sequence to find the optimal site.
- See any changes in the translated protein sequence as you design your primer.
Quicktest Primer shares the same reaction conditions as all the other primer/probe tools. So regardless of which tool you use you will see consistent results.
Give it a try.

Comments
4 responses to “QuickTest Primer: Designing Primers in MacVector”
Fabulous fabulous addition to software. 2 things though.. There appears to be an issue with being able to paste a tag sequence to the primer you are looking at. If the tag seq is actually (character wise) longer than the window you can not paste the seq. You can however type the seq in and still have the software analyze the complete primer/ tag. The other thing and this may be doable but I have not figured out how to accomplish this yet, is to easily look at a reverse primer and add a 3′ tag to it. The only way I have found to do this is to compliment/reverse the actual seq in the file and then look at the reverse primer. If this is not done I can not figure out a way to appropriately add a 3’tag to the reverse primer
Follow up. Sorry still waking up. I am actually referring to adding 5′ tags not 3′ as i initially stated in the post.
Glad you like this tool.
The pasting issue is likely a bug and I’ve entered it into our bug database. So it will be fixed in a future release. Either a minor update or in 12.7 which is in development at the moment.
For the second issue you should just be able to treat it as the forward primer. If you enter a reverse primer (i.e. the 5′ to 3′ sequence of the reverse primer on the complementary strand) then the template sequence will be “flipped” automatically so that you can just type/paste the 5′ tag into it. Do I understand your question correctly?
It is used for test the prime. it play the important role for testing.QuickTest Primer is linked to the active nucleic acid sequence window. If you select a different nucleic acid sequence window, then MacVector will recalculate the primer properties.Thanks for provide good knowledge.