Following on from my last post, and Nick’s comment that if you already have a primer collection, its not really practical to re-type them all into MacVector, one-by-one, here’s a useful tip to simplify that process.
We have a utility called PrimerConverter.app that you can download from our website. You do need to be using OS X 10.5 or later for this to run. With most web browsers, the file will automatically “unzip” when you download it. While you can run PrimerConverter.app from any location on your computer, you might want to move it to your /Applications/ folder where you should ideally keep all of your executable programs.
Before running PrimerConverter, you’ll need to get your data into a suitable format and that usually means getting your data into Excel. PrimerConverter.app can only read data from “comma separated variables” (.csv) files. Specifically it wants to see the data as a text file of one or more lines with the format;
(name), (sequence) [, (optional comment)]
e.g.
My Primer, AGCTAGTCGGGCTTTAGCTCCC, my comment
The easiest way to accomplish this is to first organize your primers in Excel so that they are laid out with the primer name in the first column, the sequence in the second column and any comments you might want included in the third column;
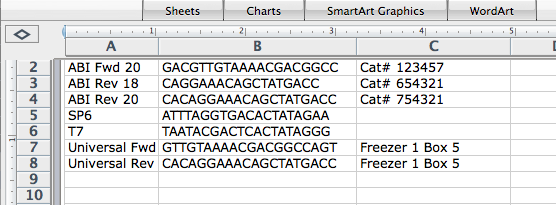
Then choose File | Save As… and select Comma Separated Values (.csv) from the Format menu and save the data to your hard drive. Now start PrimerConverter, click on the Import csv file button and select your saved csv file;
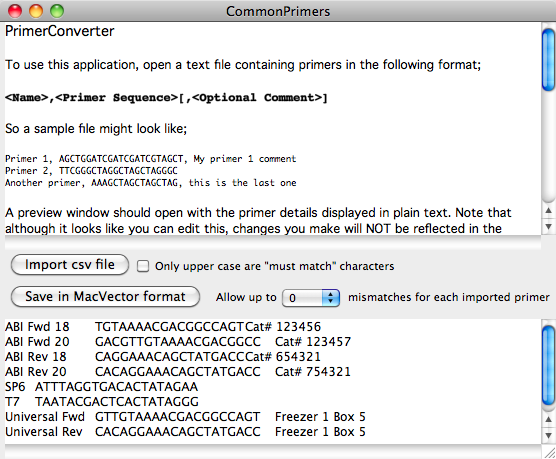
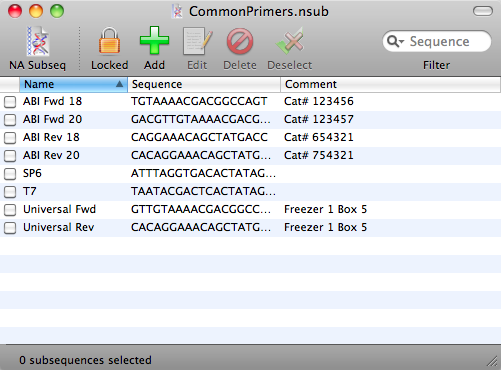
The lower text pane shows a simplified text representation of the data found in the file. If all looks OK, click on the Save in MacVector format button. If you open the saved file in MacVector, you should see that all of your primers now appear in a .nsub Nucleic Acid Subsequence window where you can use it for searches as I described in the last tip;
There are a couple of other useful tips you should bear in mind;
1) The Allow up to X mismatches… popup menu in PrimerConverter can be used to set the number of mismatches permitted for every primer in the file. I usually set this to 2 or 3 so that close matches or SNP mutagenesis sites will be picked up in the search.
2) You can also indicate residues that should always be matched by putting them in a different case. If you normally save your primers in upper case, then if you introduce some lower case residues
AAGCTTAGCTTCCCAGCATagctg
PrimerConverter will create a file where the lower case residues are treated as “must match” residues. You can also do the opposite and have most of the residues in lower case so that PrimerConverter will treat only the upper case characters as the “must match” residues. PrimerConverter assumes that the the least common case is the one marking the “must match” residues. You can override this by selecting the Only upper case are “must match” characters if you want more than half the characters to me “must match” residues.
This is an article in a long running series of tips to help you get the most out of MacVector. If you want to get notified every time a new tip gets published, follow us @MacVector on twitter (or check the feed for the hashtag #101MacVectorTips) or like us on Facebook.
