MacVector’s Subsequence tools allows you to search for motifs in both protein and DNA sequences. As well as a library of existing subsequence files, such as promotors and transcription factor binding sites, you can keep a library of your own subsequence matches. Subsequences libraries are multiple patterns kept in a single file. A search will look for matches to all subsequences in that file.
We recently had an interesting and tricky question on how to search for a protein motif where one of the amino acids was one of four different residues and the second and/or third amino acids were one of two amino acids.
Looking for ambiguous residues is relatively easy. You just surround the amino acids with parentheses and it will match one of those. For example (MV) would match either methionine or valine at that position.
However, the second part of the motif is trickier. Whereas MacVector’s Subsequence search tool can have multiple parts and you can have AND or OR, it does not accept AND/OR logic. However, you can use that OR logic to have two parts. Here’s how this was done.
Our example peptide/motif we are looking for has ten amino acids. The amino acids are as follows:
- The first position is one of five residues: arginine, lysine, aspartic acid or glutamic acid (RKDE).
- The second and third positions are where one or both are tryptophan, tyrosine or methionine (WYM).
- A string of any six amino acids.
- The tenth position can be alanine, isoleucine, leucine, methionine, phenylalanine, valine, proline or glycine (AILMFVPG).
So let’s take that motif position by position and build our subsequence.
Position 1 – (RKDE) – would match any amino acid of those four.
Positions 2 and 3 – You cannot specify that “one or both” can be a match. But you can specify that one or the other will match by using two parts with OR to match. Then Xaa = X will match any amino acid.
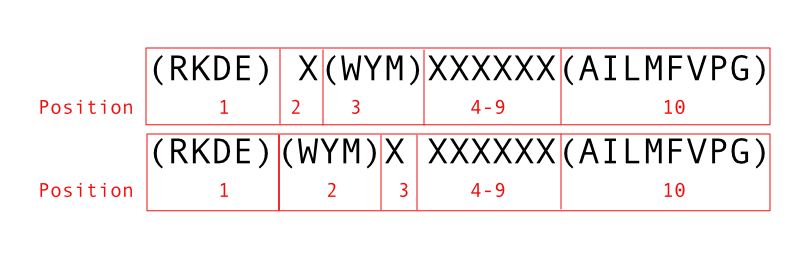
(RKDE)(WYM)X would match any amino acid at the third position.
(RKDE)X(WYM) would match any amino acid at the second position.
Positions 4 to 9 – Then you can use X for the rest. So:
- (RKDE)(WYM)XXXXXXX
- (RKDE)X(WYM)XXXXXX
position 10 will be (AILMFVPG).
So our full set of matches will be:
Here’s how this can be entered in the Subsequence Editor:
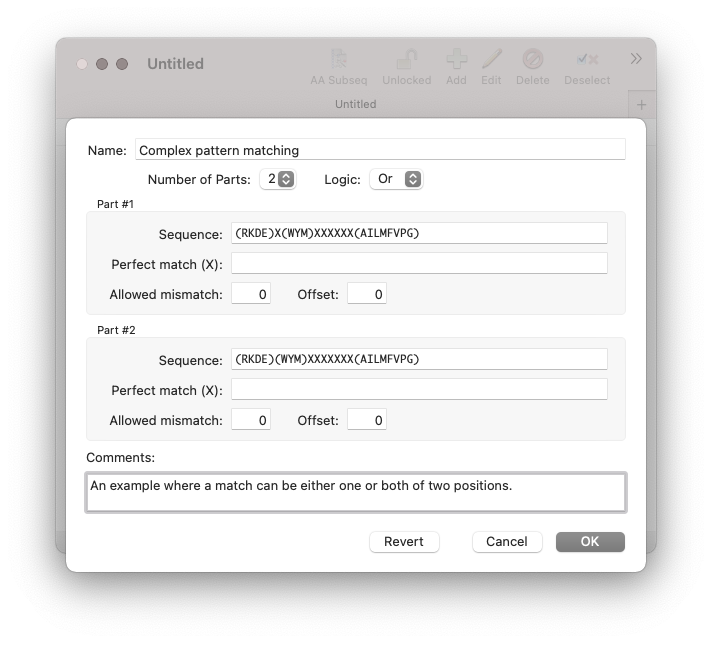
The Editing Subsequences help topic covers this.
