MacVector’s Assembly Project manager helps you organize multiple sequencing datasets, multiple reference sequences and repeated assemblies. You can store multiple assembly jobs in a single Assembly Project and directly compare multiple runs of the same dataset to determine the best assembly parameters. You can also compare different sequencing datasets assembled against the same reference sequence using the Coverage Tab.
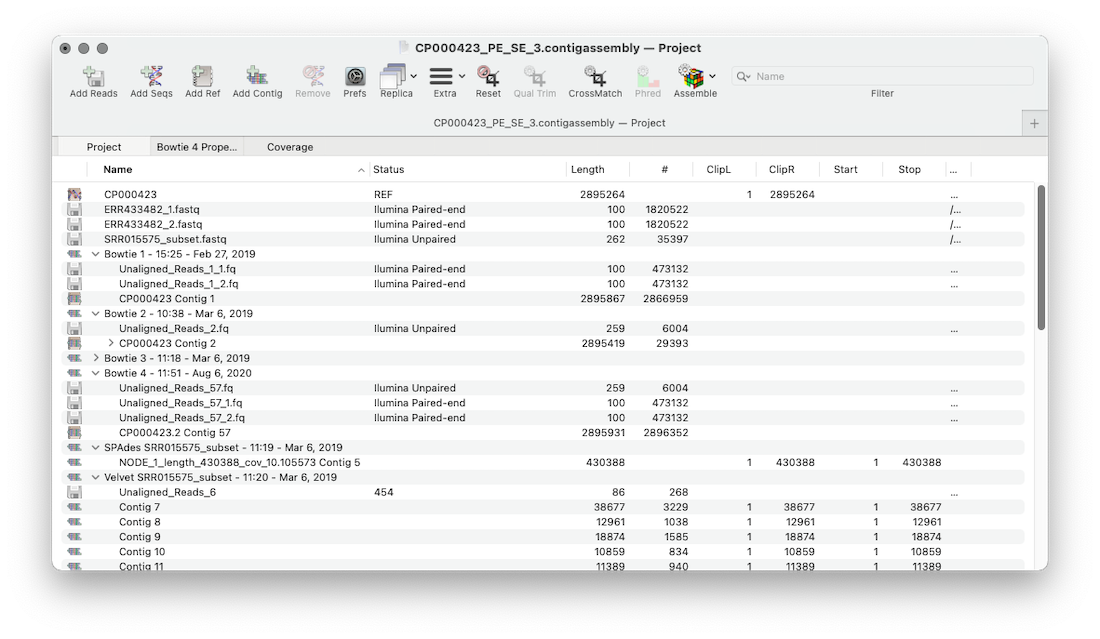
- Every run of an assembly algorithm produces a single Job Object. This is named after the algorithm used and a timestamp. For de novo assemblies the read dataset filename is also added.
- Job Objects include links to the original read datasets, unaligned reads, reference sequence(s), reference contigs (including child contigs) and de novo contigs.
- To view the properties of an individual Job Object click the 2nd tab and using the JOBS toolbar button select a job. Properties include non default parameters, read data files, statistics about the assembly including number of contigs, number of reads, N50 and more.
- To view individual objects within a Job Object click the disclosure button (triangle).
- To view contigs double click on the contig object within a Job Object.
- To view coverage depth open a contig within a Job Object, then click on the Map tab of an assembly object to see read depth visualization.
- To view expression level data click on the Coverage tab of a contig object for per gene coverage data (RPKM & TPM).
- Full export options are still available to export contigs or reads as FASTA/FASTQ files.
- New assembly jobs can be run directly on unaligned reads objects or contig objects.
