Tag: annotation
-
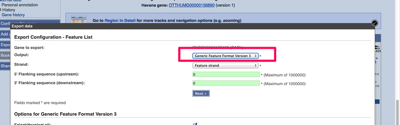
Importing sequences from ENSEMBL
There’s a few different ways to import annotation from the ENSEMBL database browser, as well as other databases. Using Genbank The easiest way to export from ENSEMBL and keep all annotation is to use the Genbank format. The default format will be FASTA which has no annotation. With Genbank all the annotation is stored in…
-
101 things you (maybe) didn’t know about MacVector: #30 – Submitting Sequences To GenBank using Sequin
Note: while preparing this blog post we discovered a bug in MacVector 12.7.4 that prevents submission using the exact steps shown here. Be sure you are using MacVector 12.7.5 or later which has the bug fixed. If you are using an earlier version, send an e-mail to support@macvector.com and we’ll send you the details of…
-
101 things you (maybe) didn’t know about MacVector: #26 – Creating Features Automatically
Following on from my recent posts on manual and semi-manual creation of features, the next approach I want to discuss is a fully automated function for creating features. How often do you get sent plain sequences that have no features annotated, even though you know your favorite gene is on there, along with a few…
-
101 things you (maybe) didn’t know about MacVector: #25 – Creating Features from Analysis Results
In my last post I described how you can quickly create and annotate features onto a DNA sequence, although the post was primarily aimed at users who are new to MacVector. In this post I’ll take a look at how you can quickly and easily annotate a DNA sequence with features based on the results…
-
101 things you (maybe) didn’t know about MacVector: #24 – Creating Features
This is a tip primarily aimed at new users of MacVector, but may be of interest to anyone who wants better understand the way MacVector handles features. MacVector can create wonderfully detailed graphical maps of a sequence, showing all the points of interest, restriction sites, open reading frames etc. However, each item to be displayed…
-
101 things you (maybe) didn’t know about MacVector: #12 – Displaying Segmented Features
While most features you might encounter in DNA files have a simple start and stop location on the sequence, some features are segmented. For example, the coding sequence of a protein encoding reading frame containing introns is represented by a segmented CDS feature on the genomic DNA. MacVector has always understood that the individual segments…
-
101 things you (maybe) didn’t know about MacVector: #2 – Option-click to open the Default Symbol editor
If you download sequences from BLAST or Entrez, or import GenBank or EMBL files with lots of features, you may wish that the features (genes, CDS, promoters etc) showed up differently in the Map tab, or maybe your favorite features don’t show up at all unless you turn them on using the Graphics Palette. Its…
-
Importing features from a Genome Browser
MacVector’s Import Features tool allows you to import annotation from many Genome Browsers (e.g. Ensemble, UCSC, etc). MacVector can annotate an empty or annotated sequence. BED, GFF, GTF, and GFF3 formats GFF, GTF, GFF3 & BED files are all file formats that are used to store annotation (features) generally without containing any sequence. Although it…
-
MacVector 12: Drag and Drop annotation of your sequences
This is in a series of blog posts on interesting highlights of new features added to MacVector 12 If you click and drag a result from a results map to a sequence window it will be annotated as a feature. For example dragging a primer from a primer results map will annotate that primer as…
-
MacVector 12: Annotating the sequence in the Editor View.
This is in a series of blog posts on interesting highlights of new features added to MacVector 12 In the last survey a popular request was to be able to change the case, alter the colour, or otherwise being able to annotate sequence directly in the Editor view. You’ve been able to use the Map…
