Tag: weeklytip
-
How to find one-out Restriction Sites that will change a protein coding sequence
If you are interested in making changes to a protein sequence, it is often useful to make a change to the coding DNA that will create a restriction enzyme site. Conversely, there may be times that you would like to create a site without affecting a protein coding region. You can accomplish this using the…
-
Use Database | Auto-Annotate Sequence to annotate prokaryotic genomes
The continuing advances in Next Generation Sequencing have made it relatively low cost to sequence prokaryotic genomes. Many scientists are embarking on large projects to sequence multiple related genomes. These might be clinical isolates of the same species exhibiting different pathogenetic properties, environmental isolates from different sites, or a study over time of the changes…
-
Automatically displaying open reading frames with MacVector’s Scan For Open Reading Frames tool
As well as Scan for.. Missing Features that shows annotation on your sequences, you may have noticed extra CDS features annotated to your sequences. These are from the Scan for.. Open Reading Frames tool that automatically scans every DNA sequence window for open reading frames and displays the results in the Map tab. It’s very useful,…
-
Pasting tabular data from MacVector into Microsoft Excel
There are a number of MacVector analyses that generate tabular text output. Examples include the Protein Analysis Toolbox List output, the Raw Data tab of the ABI chromatogram document window, the Matrix tab of the multiple sequence alignment document windows and the Coverage tab of the Bowtie contig editor. Each of these is actually composed…
-
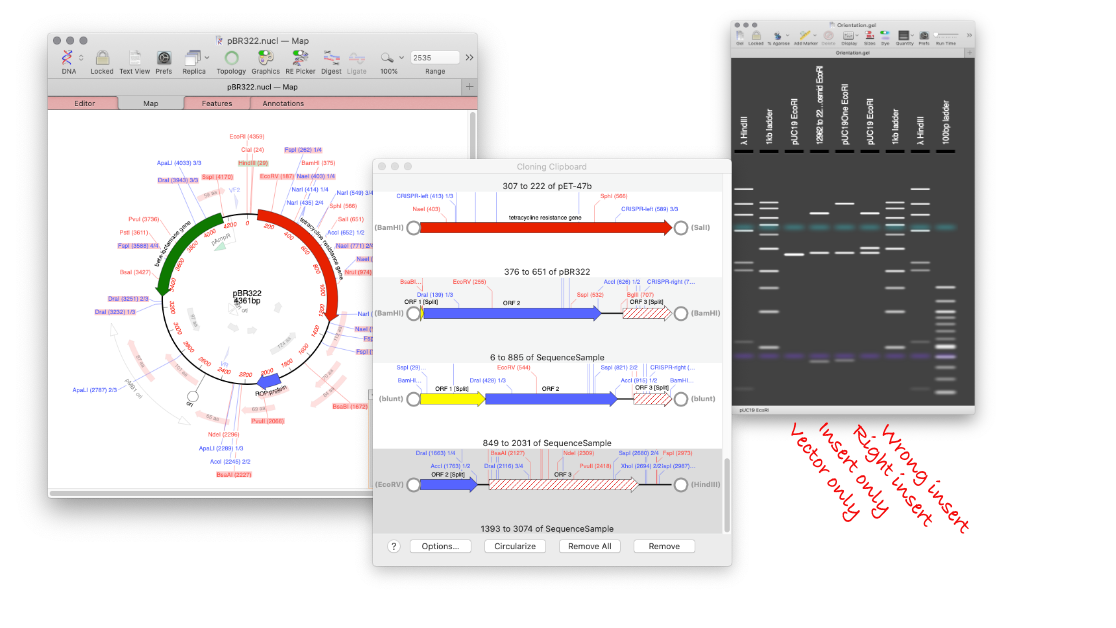
Using MacVector’s Agarose Gel tool to design a digest to screen minipreps after a ligation.
How to design a digest to screen minipreps after a ligation.
-
RNASeq Expression Analysis with MacVector and Assembler
If you have the Assembler module, MacVector can align millions of NGS reads from RNASeq experiments against large genomes and generate a coverage table displaying the relative expression levels of every gene in a genome. The key to this functionality is that you must have a reference genome with genes annotated as CDS or gene…
-
How to split large fastq files for more manageable assemblies
We’ve previously discussed how important it can be to make sure you are using the appropriate number of fastq reads from an NGS experiment to ensure you obtain the results you are looking for. Using too many reads can confuse algorithms with the massive coverage increasing mis-assemblies due to background errors in the reads. In…
-
Balancing Velvet KMER and coverage
The Velvet assembly algorithm in MacVector is blazingly fast and generates excellent assemblies. However, you do have to be careful when assembling NGS data to be sure that the parameters you submit are appropriate for the data you are assembling in order to get optimal results. By far the most important parameter is the KMER…
-
Use a right-click in the Editor tab to see if your contig can be circularized
MacVector 16 incorporates no less than THREE different de novo assemblers, phrap, velvet and SPAdes. While all are great assemblers, with each having their own specific advantages, none of them will generate a circular sequence from input reads. However, MacVector 16 also includes a new feature to help you with this. If you are assembling…
