Generating sequencing data is cheaper than it has ever been. However, with this increase in data has come a problem with easy analysis. Assembling 20 reads for your site directed mutagenesis project is easy. Why should dealing with 20 million reads of your bacterial genome be any harder?
In our Summer newsletter we talk about sequence assembly on the desktop with MacVector. We hope you find this useful and informative.
Subscribe to our newsletter and receive this every few months.
The MacVector team.
Assembler, the sequence assembly plugin for MacVector, has always been designed to be simple to use with easy visualization of your data. In our latest release, Velvet has joined the existing tools Phrap, for de novo assembly of Sanger reads, and Bowtie, for mapping reads to a reference. This gives the user great flexibility for assembling sequencing data in a single application.
De novo assembly
Velvet is a short read aligner that excels at de novo assembly of short sequencing reads. It was developed by Daniel Zerbino and Ewan Birney at the EBI. Velvet is ideal for assembling bacterial genomes on even quite modestly powered desktop or laptop Macs. Hybrid assembly is particularly straightforward and Velvet takes advantage of paired reads to scaffold between contigs.
Creating a de novo assembly
- Choose File | New | Assembly Project.
- Click on the Add Reads tool bar button.
- Click Analyze | Velvet.
For the first run use the defaults but ensure that the kmer value is lower than than the reads and make sure that paired reads is toggled appropriately.
Read mapping
Bowtie is a very fast and memory efficient aligner for mapping short sequencing reads against a reference sequence. Although it’s an ungapped assembler, what it loses in accuracy it makes up for in speed. You do not need a 32GB 8 core Mac Pro to assemble your data. Bowtie maps reads against reference sequences at a very rapid rate.
Multiple reference sequences
Mapping reads against one or more reference sequences
- Choose File | New | Assembly Project
- Click on Add Reads.
- Click on Add Ref to add the reference sequence.
- Choose Analyze | Bowtie.
Sanger sequence assembly
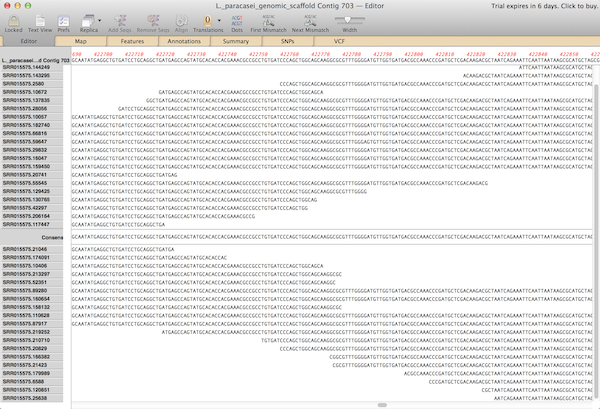
MacVector Assembler assembles Sanger trace files using Phrap. Phrap excels at producing good quality contigs using quality information produced by basecalling the reads with Phred.
Creating an Assembly
- Choose File | New | Assembly Project.
- Click on Add Reads to add your trace files.
- Choose Analyze | Phred to basecall your traces.
- Choose Analyze | Phrap to assemble the traces.
Graphical results
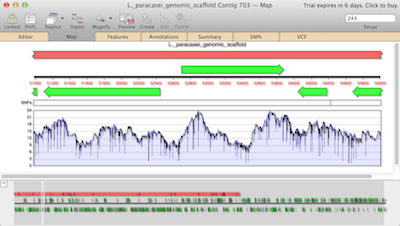
The resulting contigs from all three tools are shown at both sequence level and an overview showing the coverage map. You can zoom down to residue level to see the consensus sequence. The coverage map allows you to see areas that need more work. For example, a region that would benefit from some Sanger sequencing for hybrid sequencing. SNPs, calculated by vcftools and shown in the VCF tab, are also represented in the graphical map.
Paired end reads
Paired end reads are created by sequencing both ends of a DNA molecule of known length. Assembly and orientation of the two reads is thus less complicated.
Variant calling
All assemblies show a SNP report showing probable and possible SNPs in the consensus. Reference contigs and Velvet produced contigs also show SNPs in a VCF report.
N50
N50 is a widely used and reliable quality statistic of your assembly. The higher, the better. It is the length of the shortest contig where the sum of all longer contigs is equal to 50% of the sum of all contigs.
Summary
Assembler does all the difficult work for you. You do not have to be familiar with the command line and the myriad options for each step when creating your assembly. Just add the reads and press ALIGN. Neither do you do not need to download and install lots of software plugins (or update them with new releases). MacVector and Assembler contains all you need installed and ready to run.
Instead of sending your millions of reads away to be assembled or delving into complicated software tools you’ll be able to map millions of NGS reads to multi megabase reference sequences, or create de novo assemblies from them directly on your desktop.
Give it a try. Download the trial today!*
*If you have a license of MacVector, then a 21 day trial license will allow you to evaluate Assembler.
