Velvet is a short read aligner that works very well on a wide variety of reads. Velvet excels at de novo assembly of sequencing reads from second and newer generation sequencers.
In our latest release, MacVector 13, we’ve added Velvet to Assembler. This joins the existing tools, Phrap for Sanger sequencing reads and Bowtie for reference assembly if short reads. Assembler’s toolkit gives the user great flexibility for assembling sequencing data in a single application.
Velvet was developed by Daniel Zerbino and Ewan Birney at the EBI in Hinxton (Just outside Cambridge, UK). Amongst a large range of short read aligners available it’s widely recognised as being one of the best.
Velvet is ideal for assembling Illumina sequencing reads of bacterial genomes on a mid range Mac. With paired read data it produces very good contigs. For large datasets, especially ones with longer reads, it does use a lot of RAM. However, for bacterial genome size projects of most types of sequencing data it works very well on even quite modestly powered desktop Macs.
The nice aspect of the interface in Assembler is that all the difficult work is done for you. You do not have to be familiar with the command line and the myriad options of the application. Normally running Velvet includes preparing the project and running at least two command line tools (velvetg and velveth) to prepare your reads and then assemble them. All you need to do is import your reads in fastq format, and click run. Assembler will build the indexes needed then run the assembly.
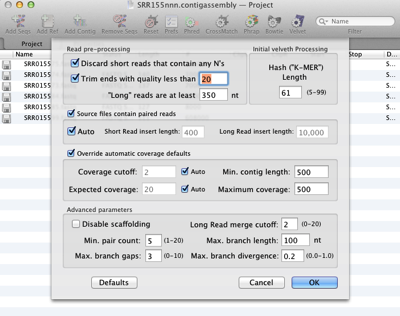
If your reads are paired end then just check the appropriate box and Assembler will do the rest for you.
Plus other than installing MacVector itself, you do not need to download and install lots of software plugins (or update them with new releases). MacVector and Assembler contains all you need readily installed to use.
A simplified explanation of the Velvet works is that it first breaks up all the reads into kmers (words) of a specified length. The kmer value must be shorter than your reads and an odd number (to minimise palindromic matches). Shorter kmers will produce longer contigs at the risk of less accuracy due to a higher probability of spurious matches, whereas longer kmers will produce more accurate contigs as the longer overlaps between kmers will be more specific. It then constructs a De Bruijn graph from the reads and uses matches between nodes on the graph to produce links between kmers and so construct the contigs. This process is repeated using reverse complemented kmers.
Hybrid assembly is particularly straightforward with Velvet. Coverage depth is not always consistent over a data set. Using longer reads sequenced really does help produce longer contigs. Long reads are treated differently to short reads and there is a different set of parameters for the longer reads. Just set the upper limit of the read length of your short read dataset and MacVector will determine which files contain your long reads and treat them accordingly.
Velvet will join two contigs as long as you have a certain number of paired reads spanning the distance between the contigs. Even though there are no reads between these two contigs. This technique is called scaffolding and really helps extend contig length. With single reads the two contigs would be separate. However, with paired reads if each read of a pair is correctly mapped against a different contig then those two contigs must be close. The insert length (the length of the sequenced fragment) determines how distant the contigs are and Velvet fills in that distance of unknown bases as a string of N’s.
The resulting contigs are shown at both sequence level and an overview showing the coverage map. However, even with the graphical map you can zoom down to residue level to see the consensus sequence. To improve performance individual reads are only shown in the editor. The coverage map allows you to see areas that need more work. For example a region that would benefit from some Sanger sequencing for hybrid sequencing.
The summary shows statistics about your assembly including the all important N50 statistic. N50 is a reliable quality statistic of your assembly. It is the length of the shortest contig in a ranked list of all contigs, where the sum of all contigs longer than this length is equal to 50% of the sum of the lengths of all contigs in the list.
Now with Assembler you have many options for mapping and processing all your sequencing reads!
References:
Velvet: Algorithms for de novo short read assembly using de Bruijn graphs
Daniel R. Zerbino, Ewan Birney
Genome Res. 2008 May; 18(5): 821–829. doi: 10.1101/gr.074492.107
PMCID: PMC2336801