MacVector 17.5 introduced Flye for assembly of PacBio and Oxford Nanopore reads.
Flye joins Phrap, Velvet and SPAdes for de novo sequence assembly using along with Bowtie2 and Align To Reference for reference assembly.
Flye is an assembler algorithm tuned to assemble low quality long reads such as those produced by the new generation of single molecule sequencers. With typical bacterial genome assemblies it is fairly common to be able to assemble reads into a single full-length genome contig.
Because these longer reads to be very error prone, MacVector also includes an optional polishing step using Racon. Polishing is the technique of correcting sequencing errors by aligning reads against contigs produced by the first run of the assembler. Multiple rounds of polishing will keep increasing the accuracy of the resulting consensus.
It is important to tell MacVector what type of reads you are assembling before running Flye. Flye will be disabled unless your read files include at least one PacBio or Nanopore dataset.
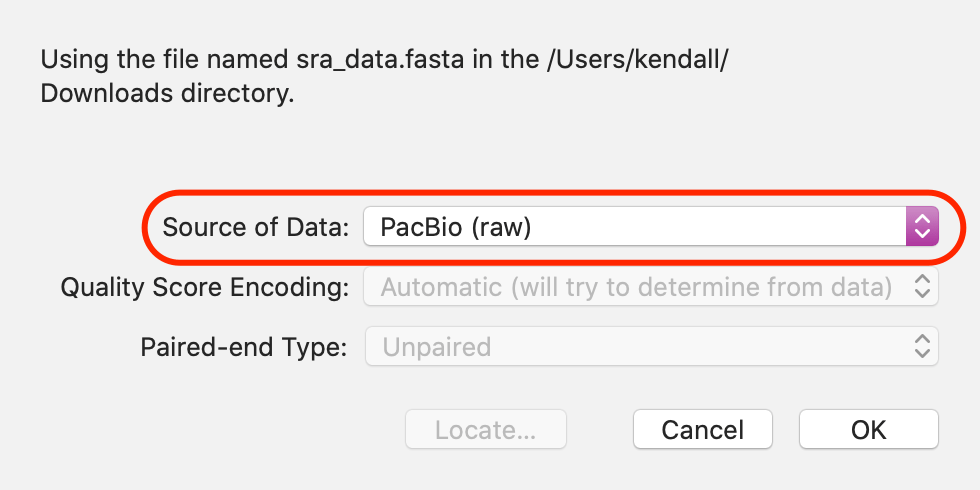
This is easily done by double-clicking on the Status item after importing the reads via the Add Reads toolbar button.
